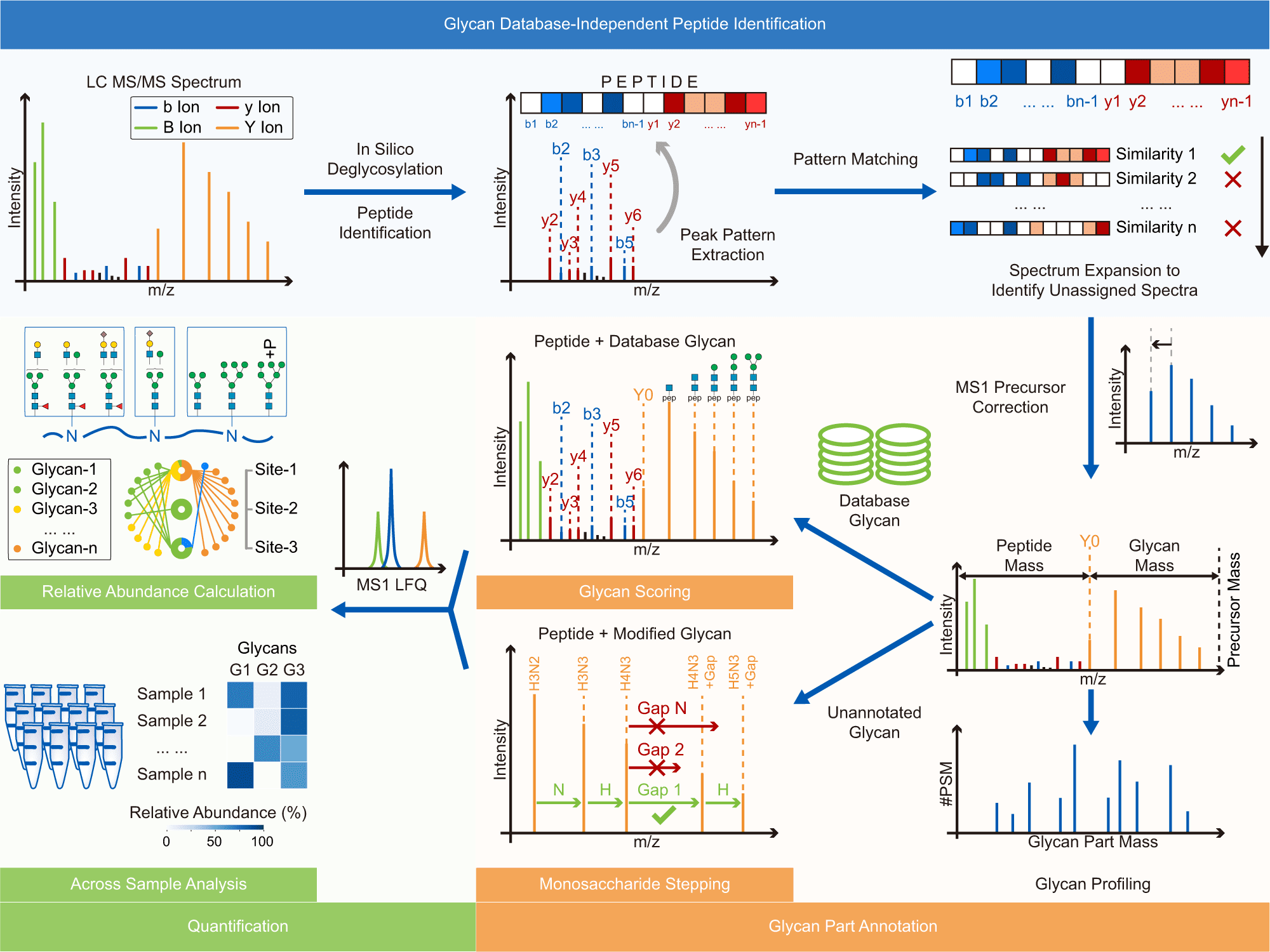
A team of scientists led by Prof. YE Mingliang from the Dalian Institute of Chemical Physics (DICP) of the Chinese Academy of Sciences (CAS) has recently created Glyco-Decipher, a new glycoproteomics software tool for the sensitive interpretation of N-glycopeptide mass spectra with high confidence. Glyco-Decipher is a promising method for enhancing glycoproteomics analysis in biological research.
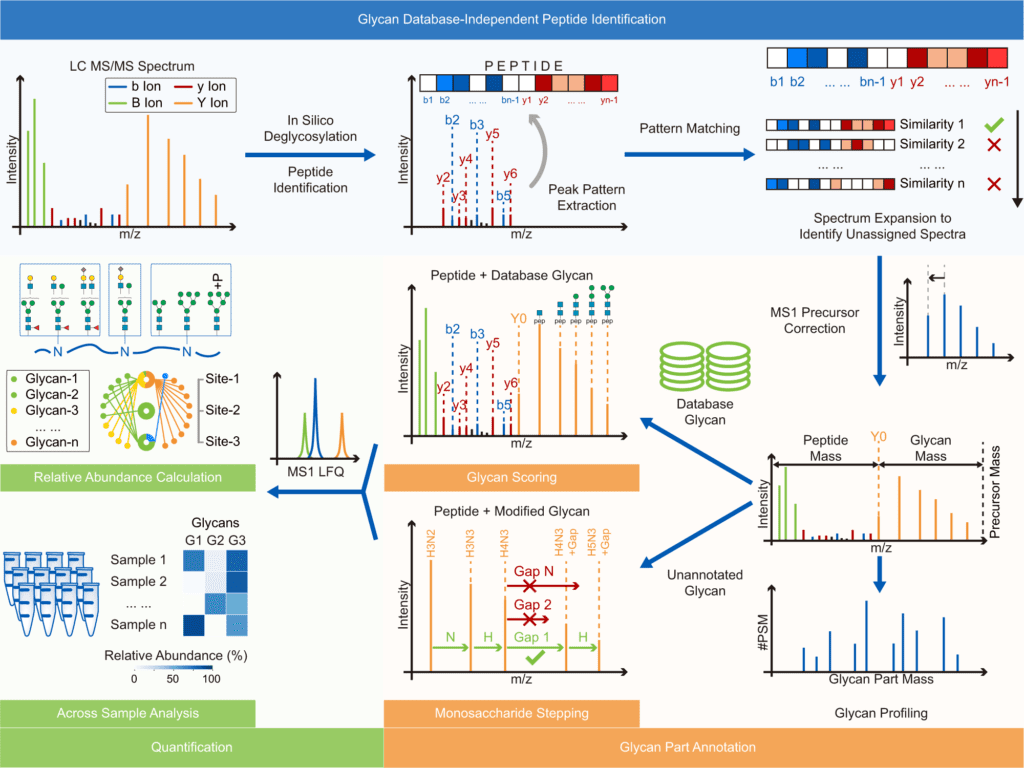
Image Source: Glyco-Decipher enables glycan database-independent peptide matching and in-depth characterization of site-specific N-glycosylation.
Glycosylation is a frequent protein post-translational modification (PTM) that is important in many biological processes, it plays a role in various biological processes, including protein folding, immunosuppression, and cancer progression. Glycosylated proteins make up a large number of illness biomarkers.
Glycosylation analysis is complicated due to the significant macro- and micro-heterogeneity of protein glycosylation. The detection of complete glycopeptides by mass spectrometry can offer information on the glycosite and the attached glycan. Many glycan database-dependent technologies for analyzing glycoproteomics data generated using tandem mass spectrometry (MS/MS) have been created in the last decade, and their efficacy is tightly connected with the adopted search variables, including the glycan database. The enormous diversity of glycans significantly raises the computational complexity of glycopeptide identification, and the interpretation of acquired glycopeptide spectra remains challenging.
Glyco-Decipher performs glycan database-independent peptide matching and takes advantage of the fragmentation pattern of shared peptide backbones in glycopeptides to improve spectrum interpretation.
According to the researchers, the connected glycans or precursor charge states did not affect the fragmentation pattern of the peptide backbone of glycopeptides. Furthermore, glycopeptide spectra with poor peptide fragmentation were interpreted using the strong similarity of the fragmentation pattern of glycopeptides with identical peptide backbones.
Glyco-Decipher surpassed other existing software tools in glycopeptide identification, according to a reanalysis of various large-scale datasets. The glycan database-free search also enables a more in-depth look at site-specific glycosylation, revealing a variety of modification moieties on glycans and as many as 164 modified glycans not found in the GlyTouCan database.
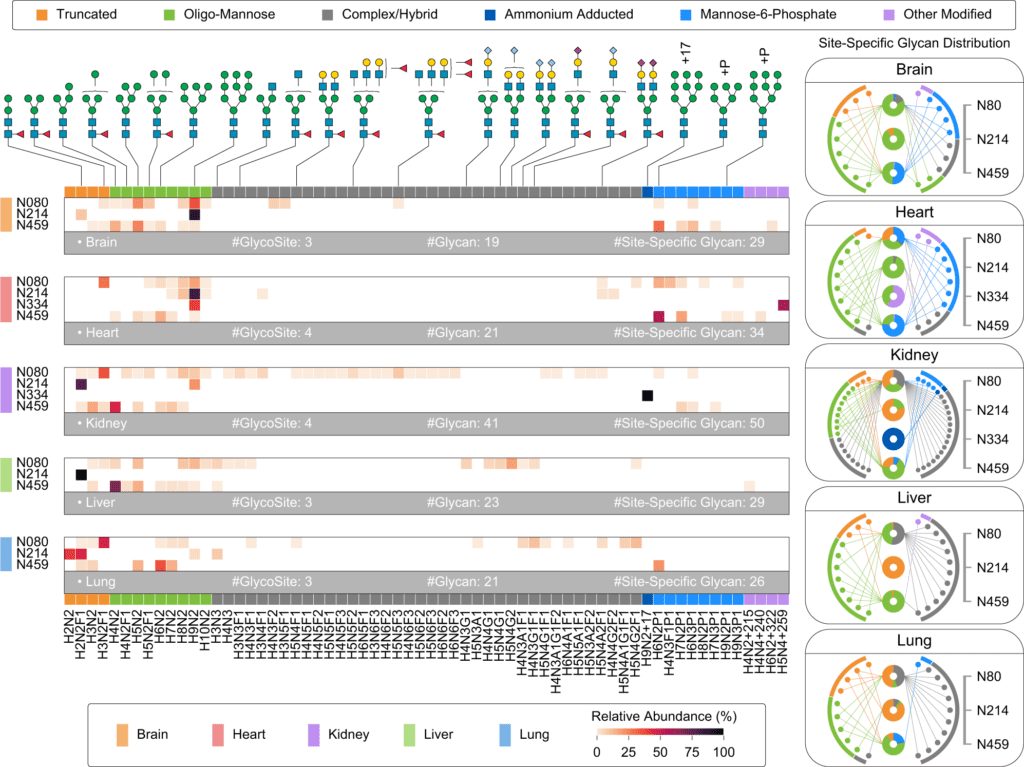
Image Source: Glyco-Decipher enables glycan database-independent peptide matching and in-depth characterization of site-specific N-glycosylation.
Furthermore, Glyco-peptide Decipher’s identification scheme was independent of glycan databases, allowing for the discovery of previously unknown glycans. Glyco-Decipher was able to uncover unexpected glycans containing modification moieties in complicated systems using the proposed monosaccharide stepping approach.
The software tool’s quantification module collected glycopeptide elution patterns, allowing for abundance analysis and functional analyses of site-specific glycans.
Indeed, more effort is required in order to improve Glyco-Decipher. To begin, Glyco-Decipher is currently designed to interpret N-glycopeptide spectra obtained from sceHCD fragmentation, with support for alternative fragmentation methods and O-glycosylation studies coming soon. Next, glycoproteomics methods such as Glyco-Decipher need to improve their detection of precursors, particularly chimeric ones. Lastly, The identification of fine glycan structures is not possible with Glyco-Decipher, and this could benefit from glycomics methods, which have been increasingly used in glycosylation analysis.
Overall, Glyco-Decipher is a glycoproteomics analysis tool that allows for database-independent profiling of N-glycans and a more thorough characterization of protein glycosylation than existing software tools. Glyco-Decipher interpreted at least 34% more glycopeptide spectra than previous methods, thanks to spectrum expansion and glycan database-independent searching.
Glyco-Decipher, which is freely available, has the potential to become a valuable tool for proteome-wide studies of glycosylation. Glyco-Decipher can be downloaded freely from: https://github.com/DICP-1809/Glyco-Decipher
Story Source: Fang, Z., Qin, H., Mao, J. et al. Glyco-Decipher enables glycan database-independent peptide matching and in-depth characterization of site-specific N-glycosylation. Nat Commun 13, 1900 (2022). https://doi.org/10.1038/s41467-022-29530-y https://english.cas.cn/newsroom/research_news/chem/202204/t20220406_303617.shtml
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.