Scientists from the University of New South Wales have discovered a complex system for adaptability in Staphylococcus aureus that has led to the identification and characterization of multi-drug resistance mechanisms as well as the development of new lines of antibiotic defense.
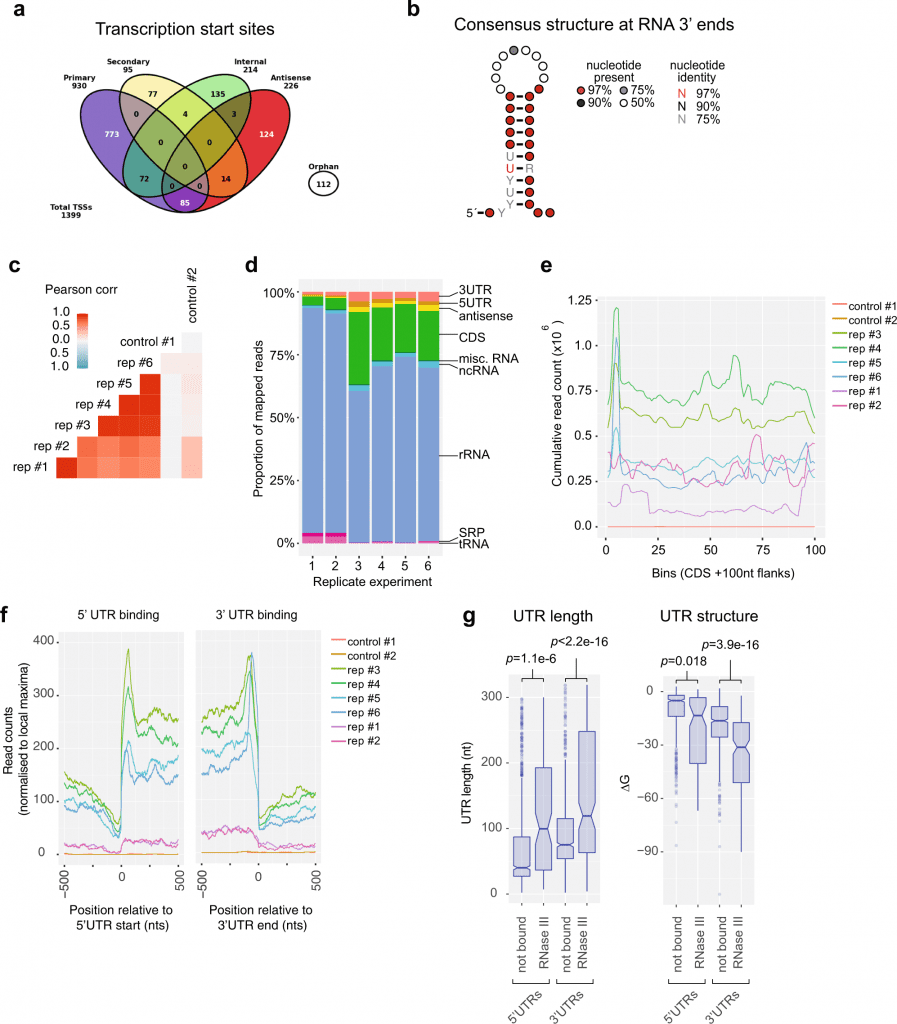
Image Source: https://doi.org/10.1038/s41467-022-31177-8
The remedy for methicillin-resistant S. aureus infections depends on the effectiveness of the last-line antibiotics such as vancomycin. The failure of last-line drugs is generally related to isolates with intermediate vancomycin resistance (termed VISA).
These isolates have developed a number of point mutations that cumulatively lessen their susceptibility to vancomycin, frequently by thickening the cell wall. In VISA isolates, variations in the expression of regulatory short RNAs have been associated with antibiotic stress; nevertheless, the roles of the majority of these RNA regulators are unclear.
A promising new tool called CLASH was used by an international group of researchers, including those from the School of Biotechnology and Biomolecular Sciences at UNSW, to capture hundreds of previously unknown mechanisms of gene regulation in a strain with antibiotic resistance i.e. S. aureus (MRSA).
The new tool uncovered 500 mechanisms that were based on the mRNA of S. aureus. These newly discovered mRNAs were influencing other genes in S. aureus through direct interactions, managing the bacteria’s genetic information and antibiotic resistance. Normally functioning only as instructions for building proteins, these mRNAs were normally controlling other genes in S. aureus.
The process that thickens the bacterial cell wall was one of the RNAs discovered; this modification is frequently observed in clinical strains of MRSA that have antibiotic resistance to the last-line drugs, and it may help uncover novel targets for antibiotic therapy. The findings of the research were reported in the journal Nature Communications.
The surprising findings
According to Jai Tree, co-author and Associate Professor, only three other mRNAs had previously been demonstrated to control bacterial RNA. It’s not that common. The actual shock came from looking at our CLASH data, though. The team discovered evidence of 543 regulatory mRNA interactions in Staphylococcus aureus.
This is a modification of how we currently understand how genes are regulated in bacteria.
Due to a lack of techniques for comprehensively collecting RNA interactions, this adaptive system in S. aureus has gone unnoticed. Similar strategies in other pathogenic bacteria depend on the presence of specific proteins; these proteins don’t appear to be active in S. aureus.
The mRNAs in S. aureus are in charge of reading DNA and then translating the DNA’s information into proteins. A regulatory role is being assumed by S. aureus, which influences the rate of synthesis of other mRNAs.
According to Jai Tree, Untranslated regions, or UTRs, are a small amount of additional sequence that is transcribed from each side of a gene (DNA) when it is converted into RNA, similar to the aglets of a shoelace. The UTRs of the mRNAs in S. aureus play a regulatory function. longer
Furthermore, the researchers discovered that about one-third of the UTRs in S. aureus are 100 bases long, which is more prolonged than the average UTR length of 40 to 50 bases. This most likely introduces an entirely new level of gene regulation.
Working on the defense’s last line
The specific strain of S. aureus employed in this investigation was isolated from a patient with MRSA septicemia who received 42 days of treatment with vancomycin, the “last-line of defense” and most potent antibiotic. The researchers used CLASH to analyze this strain in order to determine how they were evolving vancomycin tolerance because S. aureus does not naturally acquire vancomycin tolerance from other S. aureus but rather through a series of mutations.
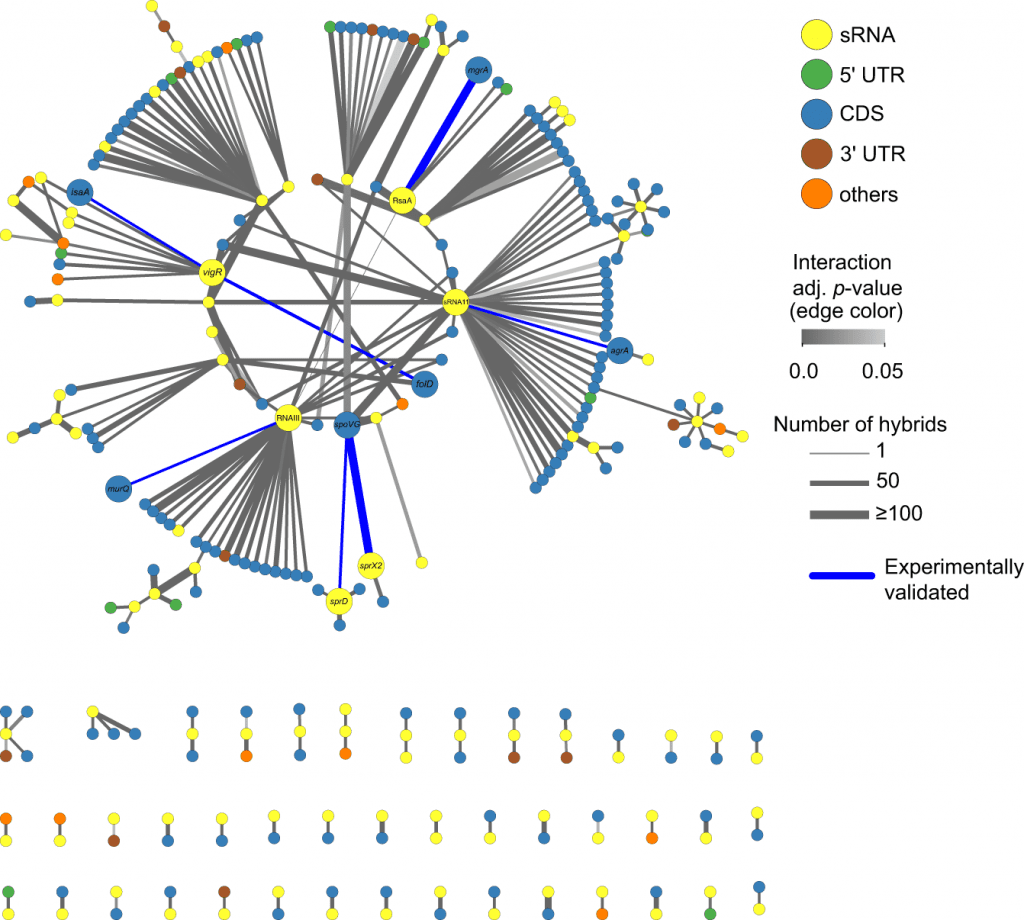
Image Source: https://doi.org/10.1038/s41467-022-31177-8
The researchers found a regulatory RNA in one of the mRNA UTRs that supports an enzyme involved in cell wall thickening. According to Prof. Jai Tree, this thickening is typical of vancomycin-tolerant S. aureus.
The researchers discovered evidence of more than 500 mRNA-mRNA interactions in S. aureus; this knowledge, made available by CLASH, enabled the researchers to assign functions to many regulatory RNAs in S. aureus frequently for the first time.
According to Prof. Jai Tree, Staphylococcus aureus, known as the “superbug,” is a serious issue in both hospital settings and the general public. Vancomycin is the preferred treatment for MRSA septicemia, an infection that enters the blood, and is only treatable with last-line antibiotics.
An antibiotic called vancomycin prevents S. aureus from forming a new cell wall. The bacterium explodes if it is unable to produce a new cell wall during division. Vancomycin resistance in S. aureus is associated with thicker cell walls, which may limit antibiotics at the site of cell wall formation.
Researchers now have the chance to take advantage of this mechanism, “re-sensitizing” resistant S. aureus to vancomycin once more. This opportunity was made possible by the discovery of one of the mechanisms underlying vancomycin tolerance in S. aureus (that previously undetected mRNAs are increasing cell wall thickness).
The use of “antisense RNA,” which can enter bacterial cells, penetrate cell walls, and bind RNAs there, has attracted increased interest. This would allow the researchers to co-administer an antisense RNA with vancomycin, making MRSA more susceptible while also killing the cell, according to Prof. Jai Tree.
Reducing antibiotic resistance in the next step
Although several mRNAs in S. aureus have been assigned roles, much work still needs to be done, especially considering the variety of ways through which S. aureus is able to tolerate vancomycin.
The following steps involve determining whether the regulatory RNA the researchers already extracted is necessary for a variety of additional clinical strains of vancomycin-tolerant S. aureus. These isolates are genetically diverse, and one of the challenges has been the variety of ways that they can develop vancomycin tolerance.
Therefore, the research team is interested in learning whether targeting the regulatory RNA might be a practical strategy for a variety of vancomycin-tolerant bacteria. Though the researchers don’t know what all the mRNAs are precisely doing, the researchers will further try to identify the important ones in the next step.
Story Source: Mediati, D.G., Wong, J.L., Gao, W. et al. RNase III-CLASH of multi-drug resistant Staphylococcus aureus reveals a regulatory mRNA 3′UTR required for intermediate vancomycin resistance. Nat Commun 13, 3558 (2022). https://doi.org/10.1038/s41467-022-31177-8 https://newsroom.unsw.edu.au/news/science-tech/new-tool-uncovers-elegant-mechanism-responsible-antibiotic-tolerance-golden-staph
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.