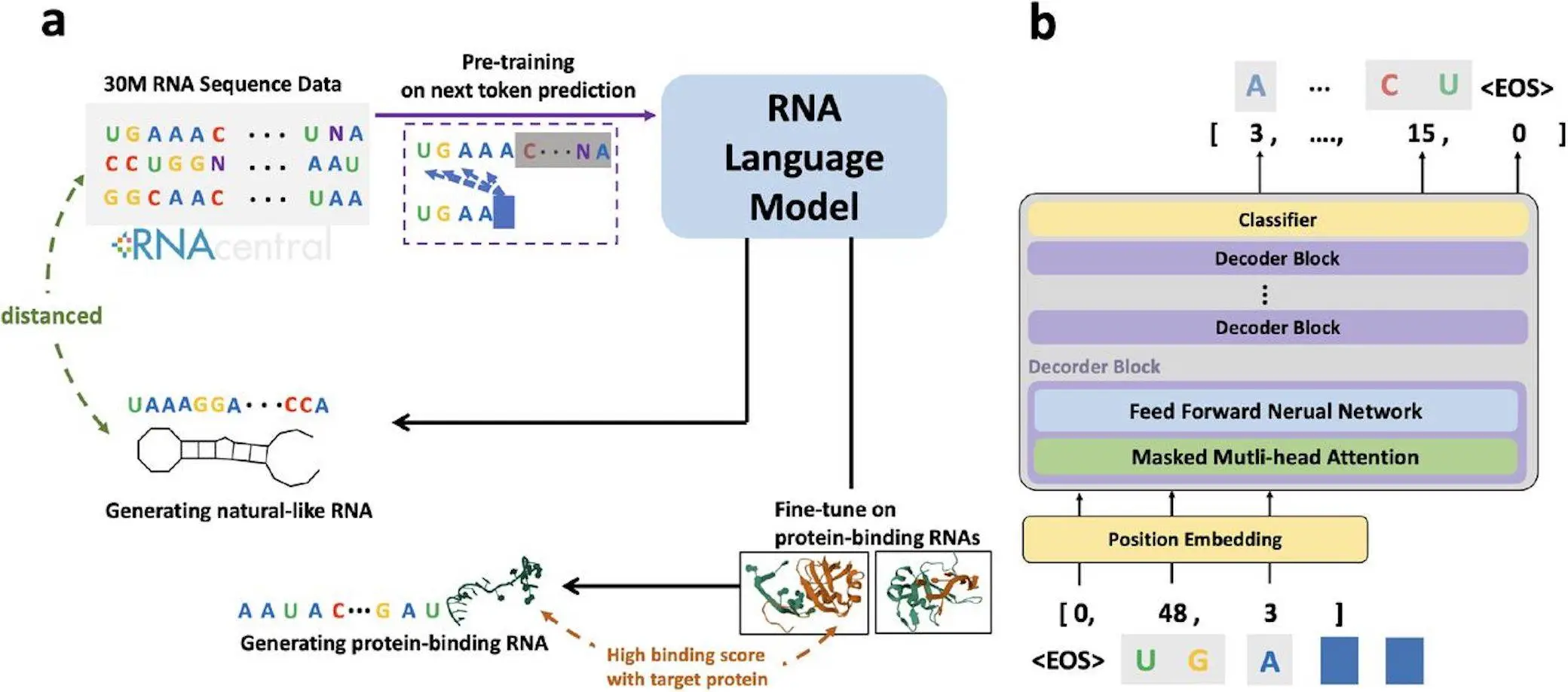
A team of researchers from the University of Tokyo developed GenerRNA, the first large-scale pre-trained AI model for automated RNA design that does not require prior structural knowledge. Inspired by advances in protein language models, GenerRNA can generate a variety of RNAs for applications such as vaccines, therapeutics, and biotechnology. This model can generate new RNA sequences with stable secondary structures that differ from existing sequences and enrich the understanding of RNA structure-function relationships. Fine-tuning allows GenerRNA to be tailored to specialized tasks, such as designing RNAs with desired binding affinity. It demonstrates the potential of artificial intelligence to accelerate RNA engineering in medicine and biotechnology.
The Need for AI-Driven RNA Design
Historically, RNA engineering has relied heavily on trial-and-error experiments, which have proven costly and inefficient. With the advent of computational techniques, the ability to design sequences with specific secondary structures has greatly advanced the field of RNA design.
However, this reliance on predetermined configurations and knowledge hinders the adaptability of RNA engineering. Fortunately, the recent success of deep generative models in generating high-quality text and images opens exciting new possibilities for automated RNA design.
Recent studies have shown promise for using them to learn protein structure and functional patterns. Similarly, models that generate RNA can effectively capture sequence-structure relationships in the context of natural language.
Introducing GenerRNA – A Transformer for RNA
To achieve AI-guided RNA design, the researchers created GenerRNA, the first large-scale RNA generation model based on the transformer-decoder architecture.
The transformer processes the input sequence and uses a self-aware mechanism to calculate the relationship between all the positions. By pre-training a large amount of unlabeled material, the characteristic patterns of a language can be learned.
GenerRNA was pre-trained on 30 million different RNA sequences from databases such as Rfam to learn the relationships between RNA “single words” (nucleotides) to gain a broad understanding of the language of RNA.
Model Architecture and Training Process
GenerRNA uses an autoregressive approach to generate sequences based on previous marker amplifications. Its 350M parameter architecture reflects the GPT-2 model processed through 24 transformer layers. Training on 18 GB of RNA sequence data minimizes the negative log-likelihood of having learned nucleotide dependencies between sequences. GenRNA requires only one week of training to learn RNA grammar and vocabulary.
Statistical Analysis Reveals Natural RNA Patterns
By examining the distribution of nucleotides, the researchers determined the optimal sampling strategy for natural RNA production. Ray searching strikes a better balance between quality and variety than greedy searching. However, random sampling best matches the frequency distribution of real RNA 3-mers and provides the most natural output. This defines the desired GenRNA decoding settings.
Assessing Structural Stability and Novelty
Evaluations of GenerRNA focused on:
- Minimum free energy (MFE) of structures as stability metric
- Novelty by homology search against known databases
The MFE distribution of the generated sequence shows significant stability at lower energy than the recombination control over length.
Homology analysis showed that 25% of the alignments with known RNAs were non-significant. Most of the aligned subsequences are still not identical, showing novelty.
Fine-Tuning Enables Targeted Protein Binding
By fine-tuning the binding of RNA to specific proteins, GenerRNA can generate high-affinity sequences. In the target in silico binding fraction, the protein exceeded the background and was close to true positive. Homology searches confirmed that the majority of associated RNAs were novel, with less than 4% identical to known examples. This demonstrates tailored optimization and creative generation.
Pre-training Drives Performance Gains
Ablation studies, minus the prior training, provide less novelty and simply repeat the training material. This highlights the important role that pre-training plays in learning a wide range of RNA functions for creative refinement.
Conclusion
GenerRNA pioneered large-scale language model RNA generation by learning complex patterns of RNA “vocabularies and grammars” from massive datasets. This is a major breakthrough in the use of artificial intelligence for versatile, automated RNA design. The refinement promises to accelerate the development of RNA therapeutics and technologies.
Continued large-scale investigation of GenerRNA function will further illuminate the potential of data-driven generative models in RNA biotechnology. By capturing sequence and structural relationships, GenerRNA provides a flexible new way to expand RNA applications.
Article source: Reference Paper | GenerRNA is freely available on: GitHub
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.