Adverse drug-drug interactions (DDIs) pose a major health risk when using drug combinations to treat complex diseases. However, predicting harmful DDIs is extremely challenging due to the intricacies of modeling interactions between drug molecules. In a new preprint posted on arXiv, scientists from Fudan University, The Chinese University of Hong Kong, and the University of California introduced an innovative deep learning framework called “DDIPrompt,” which aims to overcome two key obstacles in DDI prediction: imbalanced distribution of interaction types and scarcity of data for rare events.
The Promise and Risks of Drug Combinations
Using multiple drugs together can improve outcomes for patients with complex conditions. However, negative interactions between medications can also reduce efficacy or even modify drug structures, heightening health risks. Detecting these adverse DDI events is crucial for ensuring safe and effective treatment plans.
Graph neural networks (GNNs) have emerged as a premier technique for learning representations of drug molecules and predicting interactions. However, GNN-based DDI predictors face limitations in real-world applications.
Difficulties in Predicting Imbalanced and Rare DDI Events
Imbalanced Interaction Distributions
In collected DDI data, certain reactions occur very frequently while others rarely manifest. This skew causes models to strongly prioritize common scenarios and neglect recognizing rare but critical interactions.
Scarcity of Data on Rare Events
Furthermore, unusual DDIs often lack sufficient labeled examples to drive model training. Yet predicting these rare events is vital to safeguard patient health. Both imbalanced distributions and label scarcity hinder reliability, especially for uncommon but dangerous reactions.
Introducing DDIPrompt – A “Pre-train, Prompt” Solution
“DDIPrompt,” harnesses recent advances in language model pre-training and prompting. The core philosophy is to first pre-train an expressive model on unlabeled drug data, capturing intrinsic patterns within molecules. Then, a prompt guides the application to downstream DDI prediction using just a few examples per interaction type. This leverages innate knowledge within pre-trained weights while drastically reducing labeled data dependence.
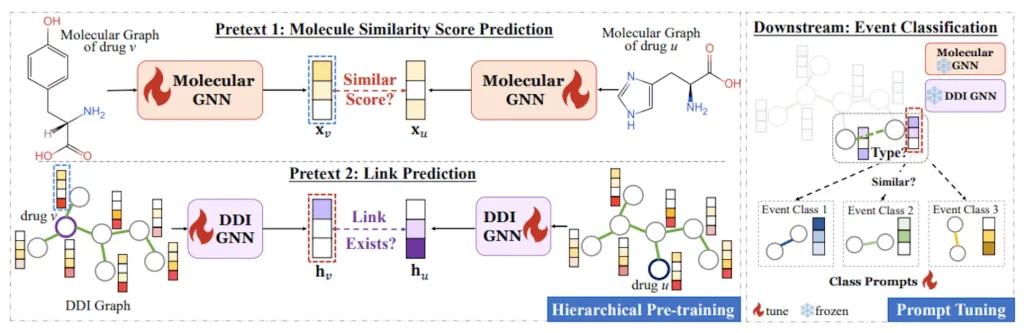
Image Source: https://doi.org/10.48550/arXiv.2402.11472
Hierarchical Pre-Training Learns Robust Drug Representations
Capturing Intra-Molecular Structures
The first stage of DDIPrompt pre-trains a GNN encoder on molecular graphs to comprehend functional groups and bond connections within individual drugs. This ensures a rich understanding of pharmacological properties.
Modeling Inter-Molecular Relations
The next phase analyzes relationships spanning across drugs based on structural and interactive similarity. Learning these inter-molecular associations reduces reliance on just DDI labels, which can be sparse or biased.
Together, hierarchical pre-training builds an unbiased, comprehensive representation of drug chemistry for downstream interaction prediction.
Prototype-Enhanced Prompting for Few-Shot Inference
Prompt Vectors as Interaction Prototypes
During inference, DDIPrompt feeds samples of each DDI type into prompt vectors that essentially serve as “prototypes” summarizing key properties. Fine-tuning these prototypes on available data adapts the model to recognize nuances of each interaction class.
Efficiently Harnessing Pre-Trained Knowledge
This prompting rapidly aligns pre-training objectives with the downstream prediction task. By exploiting patterns already within model weights, only a few examples suffice to tune prompts and extract information necessary for accurate classification.
The prototype-based prompting grants robust performance even under low-data conditions. Combined with robust pre-training, DDIPrompt finally overcomes key obstacles in reliable DDI prediction.
Experimental Results Validate Effectiveness on Benchmarks
Consistent Improvements in Prediction Accuracy
Evaluations on real-world drug interaction datasets demonstrate DDIPrompt’s superiority over leading GNN baselines and few-shot learning techniques. Impressively, F1 scores for rare events showcase remarkable 24-69% jumps over competitors.
Each framework component, including pre-training strategies and prompting, contributes notably to predictive gains. Ablation studies confirm that removing any piece degrades end-to-end performance.
Robustness Under Varying Low-Resource Scenarios
Further analysis reveals that DDIPrompt maintains strong accuracy even when utilizing just 10-20% of samples per class for final tuning. Performance scales steadily as more training data is provided.
In contrast, existing methods prove far more sensitive to limited labels. This showcases DDIPrompt’s scalability down to extremely sparse interaction data.
Conclusion
DDIPrompt sets a new state-of-the-art in adverse drug-drug interaction prediction, finally conquering the pressing issues of label imbalance and rarity through innovations in pre-training and prompting. Hierarchical learning of molecular patterns couples with prototype-based few-shot tuning to enable reliable detection even for uncommon, hazardous reactions.
Moving forward, expanding the diversity of pre-training signals has the potential to enrich model understanding and enhance adaptability to even more drug safety prediction tasks. Robust DDI detection will pave the way towards safer and more optimized use of drug combinations.
Article source: Reference Paper
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.