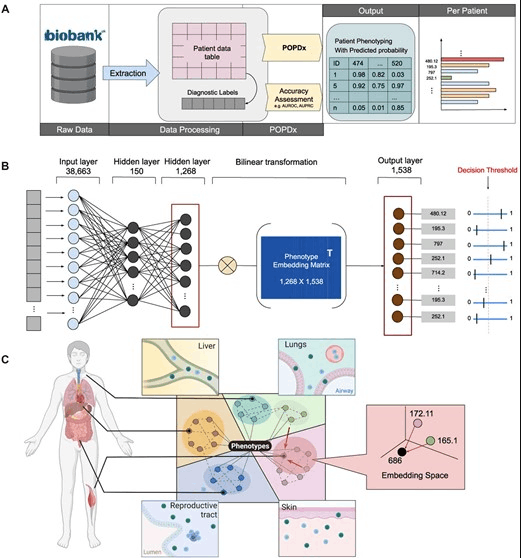
The POPDx model does not require extensive patient datasets, which makes it potentially useful for patients with rare diseases.
The POPDx framework was created to effectively identify a wide variety of phenotypes among participants in the UK Biobank study. To provide a thorough analysis, it makes use of a wide variety of patient data, including demographic details, medical histories, lifestyle factors, and genetic information. The framework has been successfully tested on the UK Biobank dataset and outperforms other comparable methods in identifying a wide range of phenotypes, both common and rare. This ground-breaking platform has the potential to increase the precision of phenotype recognition significantly and can be used with other biobank records.
In order to advance precision medicine and enhance patient care outcomes, POPDx is positioned to emerge as a significant player in the field of patient phenotyping. The results offer useful direction for scientists, medical professionals, and patients.
Researchers can examine diseases and look into how genetics and environment affect disease progression thanks to biobanks, which store genetic and medical information. Understanding different facets of the disease, such as how diet affects health and the influence of household size on COVID-19 severity, has been made possible by this research.
The usefulness of biobanks, however, depends on the quality and quantity of the data. The incompleteness of patient data is a frequent problem. For instance, even though a patient may be listed as having type II diabetes, this information might not be present in their data if they have never been admitted to the hospital. Scientists who are researching diseases and looking for potential novel discoveries using pattern recognition face a significant challenge because of this missing data.
For every patient in the UK Biobank, the researchers created a model to predict the complete set of diagnosis codes, also known as phenotype codes. Information from 500,000 UK participants, including those with rare diseases, is stored in the UK Biobank. The research team developed a model called POPDx, a machine learning framework for disease recognition, to provide probabilities of a person having specific diseases or phenotype codes.
The research findings show that POPDx outperforms current models in diagnosing both common and rare diseases, including those not included in the training data. Given that conventional deep neural network machine learning approaches frequently need a lot of training data, this represents a significant accomplishment. The research team was able to correctly identify some diseases in the test set even though those diseases were not seen in the training data because of how the method used prior knowledge from text and taxonomy. This is crucial because the medical industry does not have access to the same volume of data as larger IT firms, making it imperative to create techniques that can efficiently use this limited data and help patients with rare diseases.
Comprehensive patient analysis
The study started by taking into account the data from one of the author’s prior research work on the classification of cells. The correct cell type for each of the trial set’s cells was determined using Cell Ontology. In this study, the authors also adopted a similar strategy for diseases. The researchers believed it would be fascinating to use the Human Disease Ontology’s connections between diseases to identify diseases. This study required multiple labels, in contrast to the previous research, which was a one-vs.-all classification problems where only one cell type was identified. Patients could have multiple diseases, thus, a multi-label and multi-classification approach was used.
The POPDx model looks at a lot of patient data, including demographics, patient surveys, physical and lab test results, EHR data, and medical examinations. The number of features that earlier models could analyze was constrained by the requirement for well-organized datasets. However, the broad scope of this research makes it possible for the model to predict a wide variety of disease codes. This study aims to present a thorough profile of the UK Biobank participants, in contrast to most research focusing on a particular domain, such as heart disease.
Disease prediction with limited amount of data
Human Disease Ontology and natural language processing are used in the POPDx approach to examine connections between the patient’s data and details of their diseases. This enables the model to make probabilistic decisions. The biggest challenge for the model is dealing with diseases for which there is either little or no training data available. It’s important to note that most machine learning models need a lot of data to function, but some diseases lack the necessary data.
Despite the dearth of data, the remarkable performance of POPDx displaces the need for large data sets. For invisible and rare diseases, the research team was able to increase the AUPRC (precision metric for the model) by 218% and 151%, respectively. As a result, clinical teams looking for patients with rare diseases can now screen fewer individuals in the Biobank, increasing the chance of discovering positive cases. The ability of POPDx to identify rare diseases provides clinicians and researchers working on those diseases with a more useful starting point.
The UK Biobank has an imbalanced demographic, with 56% of its participants being women, the majority of whom are white, and an average age of 71, which is challenging. The access to healthcare, not data problems, is to blame for this lack of diversity. If a person does not have access to healthcare, there is no data on them. The model’s capacity to handle rare diseases was enhanced by the researchers’ incorporation of background knowledge regarding the connections and relationships between various diseases. This strategy might have also increased unpredictability and lessened biases. The researchers hope that more infrastructure will be built in the future to combine data from various biobanks, creating a dataset with a wider range of information.
The future of patient phenotyping
The researchers intend to conduct additional analyses of the patient data over time, looking at both the likelihood that a patient will develop a disease and the potential timing of that development. Another choice is to include information on genotype and phenotype in the model, which provides a complete understanding of diseases. The researchers are looking forward to creating inclusive models that benefit everyone, as access to data is essential for both patients and researchers.
Article Source: Reference Paper | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.