Scientists from the University of California, San Francisco, have created an inventive AI system that can create entirely new enzymes from scratch. Even though the amino acid sequences of these synthetic enzymes differ significantly from those of any naturally occurring protein, they have demonstrated remarkable efficacy in laboratory experiments. In other words, using state-of-the-art technology, researchers can produce enzymes that are just as efficient as those found in nature but have entirely different characteristics.
The field of biotechnology has recently seen some exciting advancements thanks to deep-learning language models. One such model, ProGen, can create protein sequences that perform specific functions within large protein families. This is similar to how a person can generate grammatically and semantically correct sentences on various topics using natural language.
What is ProGen?
ProGen is an AI program that utilizes advanced prediction techniques to construct artificial proteins using a sequence of amino acids. ProGen was developed in the year 2020 by Salesforce Research.
Based on prior experiences, the research team was confident that the AI technology could learn grammar, comprehend word meanings, and adhere to other crucial standards essential to producing polished and well-structured written content.
An extensive dataset of 280 million protein sequences from over 19,000 different families has been fed to the sophisticated AI model. In order to fine-tune the protein properties for even more accurate results, researchers have added additional control tags.
This cutting-edge technology has the potential to surpass directed evolution, a protein design method that earned a Nobel Prize, and is set to revolutionize the field of protein engineering. Expediting the creation of new proteins could be used for a wide scope of applications, including therapeutics and even breaking down plastic.
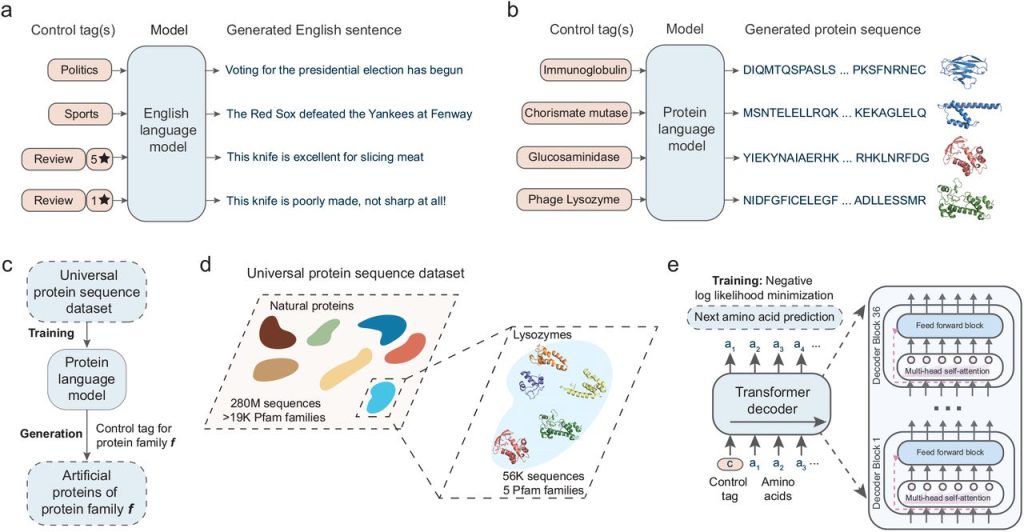
Image Source: https://doi.org/10.1101/2021.07.18.452833
How Advanced Machine Learning is Revolutionizing Protein Engineering?
Although the language model operates differently from conventional evolutionary processes, it is still learning aspects of evolution. Now it is possible to precisely control the generation of particular properties for particular uses thanks to this new technology. For instance, we can design enzymes that operate effectively in acidic environments, withstand high temperatures, and avoid interacting with other proteins.
The team developed the model using a simple approach. They first fed the machine learning model with many amino acid sequences, totaling 280 million proteins of various types. After allowing the model to process this information for a few weeks, they fine-tuned it by providing it with additional data, including 56,000 sequences from five lysozyme families and some related information about these proteins.
The research team was amazed at how quickly the model produced a massive number of sequences. They carefully handpicked 100 sequences to experiment on based on their similarity to natural proteins and how closely the AI proteins resembled the underlying amino acid “grammar” and “semantics” of real proteins.
After analyzing a collection of 100 proteins from Tierra Biosciences, the team selected five of them for further testing. The proteins’ activity within cells was examined and compared to an enzyme commonly found in chicken egg whites, known as hen egg white lysozyme (HEWL). Lysozymes, similar to HEWL, are also present in human tears, saliva, and milk and play a role in protecting against bacteria and fungi.
Two synthetic enzymes displayed an impressive ability to destroy the protective cell walls of bacteria, matching the activity of a well-known enzyme called HEWL. What’s even more remarkable is that these two enzymes have vastly different sequences, with only 18% similarity between them. Additionally, their sequences bear little resemblance to any known protein, with similarities of only 90% and 70%, respectively.
Natural proteins are incredibly delicate, and even a small mutation can render them ineffective. However, in a recent study, the team discovered that AI-generated enzymes were much more resilient. Even when they only shared 31.4% of their sequence with known natural proteins, they still displayed activity.
The AI system is truly impressive in its ability to understand how enzymes should be structured, all from analyzing raw sequence data. It was amazing to see that, when examined using X-ray crystallography, the atomic structures of the artificially created proteins matched expectations, even though the sequences were completely unique.
When a lot of data is provided to sequence-based models during training, they become incredibly skilled at understanding patterns and rules. They can figure out which words commonly appear together and how they work together to form meaning.
Protein design has countless potential applications. Consider lysozymes as an example. Despite their small size, these proteins can be combined with up to 300 amino acids, and there are 20 different types to choose from. It’s truly astounding that the model can produce functional enzymes with such ease, considering the endless options available.
Since researchers have found a way to build functional proteins from scratch, protein design is about to enter a revolutionary period. It is exciting to explore the incredible therapeutic possibilities that this ground-breaking method opens up for protein engineers.
Benefits of Using a Natural Language Modeling for Protein Generation
- Greater accuracy: Natural language models can be used to generate proteins that are likely to fold correctly and perform the desired function. Algorithms can analyze large amounts of data about protein structure and function and use it to create new proteins with a higher chance of success.
- More efficient: The protein engineering techniques used in the past were often time and resource intensive. Language modeling allows researchers to create new proteins much faster than with more conventional techniques.
- Ability to generate proteins across families: One of the most exciting features of the model ‘ProGen’ is its ability to generate functional proteins across different families, enabling scientists to design proteins with specific functions even if they don’t exist in nature.
- Potential for new therapeutic applications: Researchers can develop new proteins with therapeutic uses by being able to produce functional proteins from different families. To target particular diseases or conditions, scientists can develop proteins.
Conclusion
The research demonstrates the ability of a state-of-the-art language model, ‘ProGen’ that only trains on evolutionary sequence data and makes use of transformer technology to produce useful artificial proteins across a range of protein families. Further investigation reveals that the model has mastered the ability to modify its protein sequence approach for various families, including lysozymes, chorismate mutase, and malate dehydrogenase. The model could be used to create artificial collections of exceptionally effective proteins for ongoing research or advancements. This study suggests that using deep learning-based language models for the precise creation of new proteins may be a promising solution for various problems in biology, medicine, and the environment.
Article Source: Reference Paper | bioRxiv
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.






