A team of scientists and engineers at the University of Texas at Austin created an enzyme that could break down environmentally damaging polymers in hours to days instead of centuries.
The breakthrough discovery published in Nature could help to tackle one of the world’s most important environmental issues: how to deal with the billions of tonnes of plastic garbage stacking up in landfills and polluting natural lands and water resources. The enzyme has the power to increase recycling on a large scale, allowing large corporations to decrease their environmental effect by recovering and reusing plastics at the molecular level.
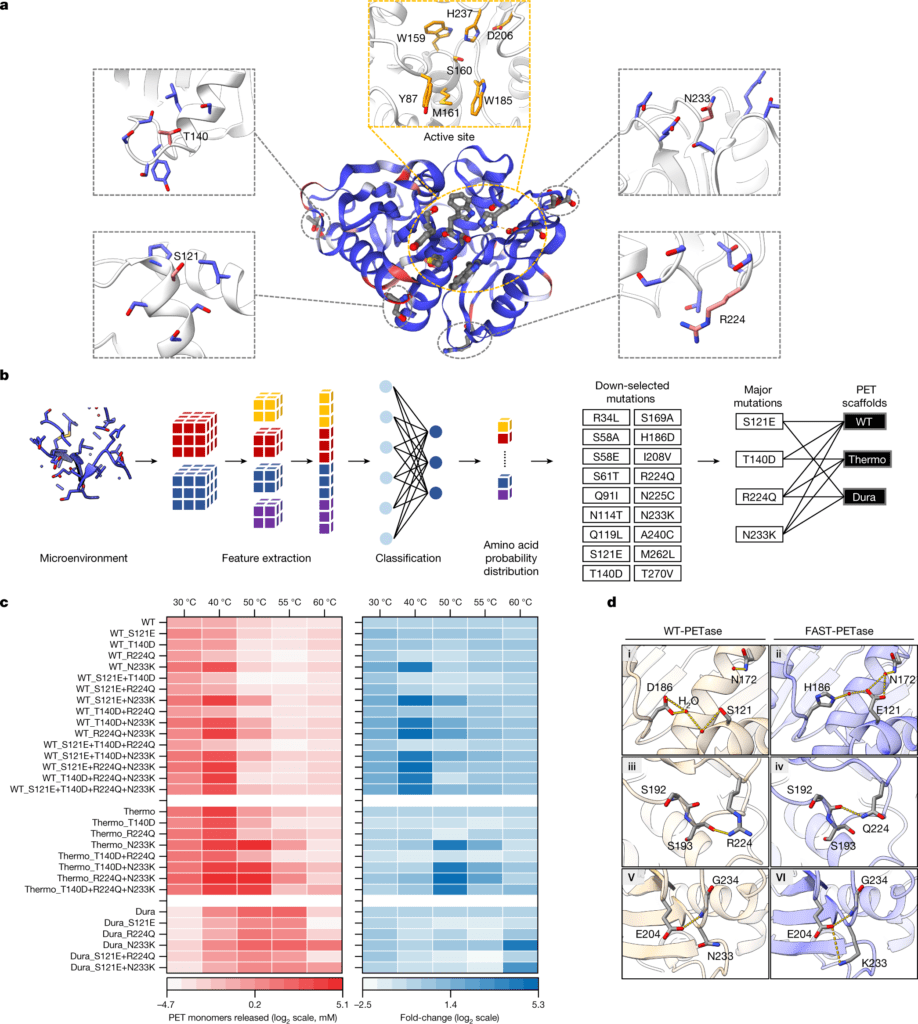
Image Source: Machine learning-aided engineering of hydrolases for PET depolymerization
The possibilities are endless across industries to leverage this leading-edge recycling process. Beyond the obvious waste management industry, this also provides corporations from every sector the opportunity to take a lead in recycling their products. Through these more sustainable enzyme approaches, we can begin to envision a true circular plastics economy.
Hal Alper, professor in the McKetta Department of Chemical Engineering at UT Austin
PET, a significant polymer present in most consumer packaging, including cookie jars, soda bottles, fruit and salad packing, and some fibers and textiles, is the subject of the research. It constitutes about 12% of all garbage over the world. Plastic waste poses an ecological challenge, and enzymatic degradation offers one potentially green and scalable route for polyesters waste recycling.
The enzyme was able to perform a “circular process” that entailed plastic breaking down into tiny bits (depolymerization) and then chemically reassembling it (repolymerization). In some cases, the plastic waste can be entirely split into monomers in a single day.
Researchers at the Cockrell School of Engineering and College of Natural Sciences applied a machine learning model to create unique mutations in PETase, a natural enzyme that allows bacteria to break down PET polymers. The model predicts which enzyme mutations would let post-consumer waste plastic to be depolymerized quickly and at lower temperatures.
The efficiency of the enzyme, which they call FAST-PETase, was confirmed by testing 51 different post-consumer plastic containers, five different polyester fibers and fabrics, and PET water bottles (functional, active, stable, and tolerant PETase).
This work really demonstrates the power of bringing together different disciplines, from synthetic biology to chemical engineering to artificial intelligence.
Andrew Ellington, Professor, Center for Systems and Synthetic Biology, led the team to develop the machine learning model
The most obvious way to reduce plastic waste is recycling. However, globally, only around 10% of all plastic has been recycled. Apart from depositing it in a landfill, the most common method of disposing of plastic is to burn it, which is costly, energy-intensive, and emits toxic gas into the atmosphere. Glycolysis, pyrolysis, and/or methanolysis are some more energy-intensive alternative industrial processes.
Biological solutions require significantly less energy. Enzyme research for plastic recycling has evolved over the last 15 years. Though, no one had been able to find out how to manufacture enzymes that could function effectively at low temperatures and be portable and economical on a huge industrial scale until now. FAST-PETase can work at temperatures as low as 50° Celsius.
The team’s next goal is to scale up enzyme production so that it may be used in industrial and environmental applications. The researchers filed a patent application for the technology and are eyeing different applications. Landfill cleanup and greening, high-waste-producing businesses are the most prominent examples. Another possible key application is environmental cleaning. The team is examining numerous strategies for delivering the enzymes to polluted locations in the field.
When considering environmental cleanup applications, you need an enzyme that can work in the environment at ambient temperature. This requirement is where our tech has a huge advantage in the future.
Hal Alper
Story Source: Lu, H., Diaz, D.J., Czarnecki, N.J. et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 604, 662–667 (2022). https://doi.org/10.1038/s41586-022-04599-z https://news.utexas.edu/2022/04/27/plastic-eating-enzyme-could-eliminate-billions-of-tons-of-landfill-waste/
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.












