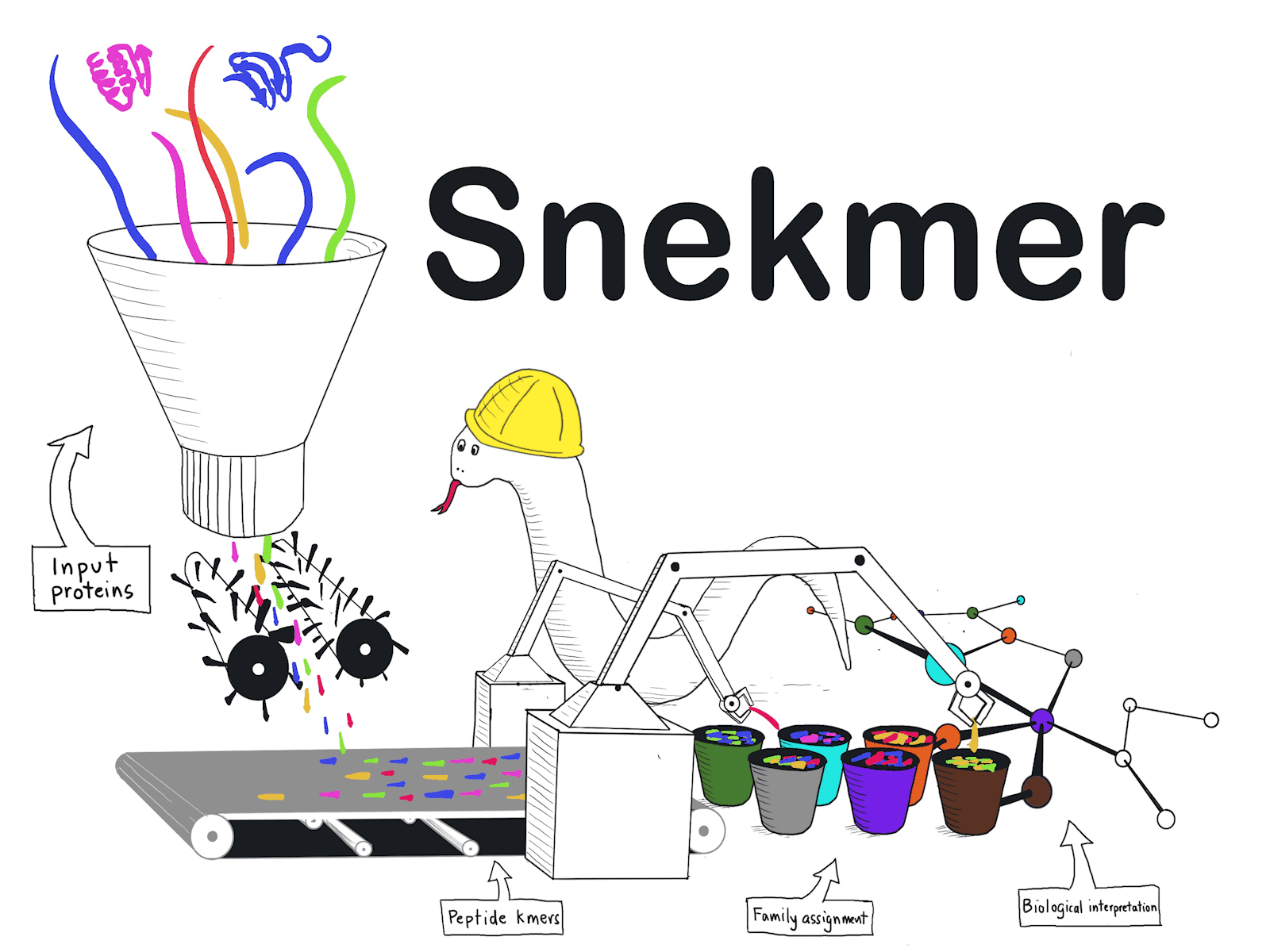
Snekmer, an innovative software developed to better understand protein function in microbes, emerged from the collaborative efforts of researchers from the Pacific Northwest National Laboratory, Baylor University, and Oregon Health & Science University. This cutting-edge tool harnesses the power of short protein sequences, known as kmers, to generate protein family models through machine learning techniques. Furthermore, Snekmer leverages the redundancy in amino acid residue properties to reduce the sequence space effectively. Users of Snekmer are granted the capability to convert protein sequences into concise alphabet kmer vectors, construct supervised classification models trained on input protein families, and classify proteins based on their functions utilizing the robust Snekmer models.
Microbes play a vital role in crucial processes on Earth, impacting the circulation of carbon, nitrogen, and various elements within elemental cycles. They also influence the onset of diseases and promote plant growth, making their presence essential in every ecosystem. While the library of microbial DNA sequences continues to expand, it lacks comprehensive biological information about proteins. To advance the development of microorganisms for sustainable bioenergy and other bioproducts, scientists require a deeper understanding of protein function and other molecular operations. Traditionally, scientists have inferred protein functions by comparing them to reference databases containing known proteins. However, this approach poses challenges and is not adaptable to large datasets.
Protein structure, function, and evolution can all be understood better, thanks to protein sequence fingerprinting. It involves the identification of conserved patterns or motifs within protein sequences that can provide valuable insights into their biological significance. Snekmer is a revolutionary pipeline that utilizes amino acid recoding (AAR) techniques to enhance the scalability and accuracy of protein sequence fingerprinting.
The Need for Protein Sequence Fingerprinting
The building blocks of life, proteins, serve a wide range of purposes in cells. Understanding their sequence patterns and motifs can unravel their functional and evolutionary relationships. Protein sequence fingerprinting techniques help identify conserved regions across diverse protein sequences, enabling researchers to infer functional similarities and evolutionary connections.
Traditional methods of protein sequence fingerprinting often face challenges in terms of scalability and accuracy. There is a need for effective algorithms that can handle large-scale analysis without sacrificing accuracy as the volume of protein sequence data continues to increase rapidly.
Enhancing Protein Analysis with Snekmer
Snekmer is an open-source Python tool that extends the AAR approach into a flexible pipeline. It offers various recoding schemes, model training, and automatic evaluation capabilities. Users can utilize Snekmer to build classification models or perform clustering analyses on protein datasets. The tool supports high-performance computing environments, allowing efficient processing of large sets of protein sequences.
Snekmer operates in two main modes: supervised and unsupervised. In the supervised mode, users can train models on input protein families by providing FASTA files containing sequences from each family. Snekmer generates AAR features, constructs feature vectors, and calculates probability scores for each kmer feature based on its representation within and outside the family. Logistic regression models are then built for each family, allowing the classification of new sequences. Snekmer performs clustering analysis in the unsupervised mode to identify similarities between protein sequences in a given FASTA file.
Key Features of Snekmer
Snekmer offers several key features that make it an exceptional tool for protein sequence fingerprinting:
- Snakemake determines dependencies between workflow steps and executes them sequentially.
- Snekmer may be utilized in high-performance computing settings like supercomputers and cloud computing clusters since it is very scalable.
- Each step in the workflow is executed as a separate job script, allowing for parallel processing of multiple input files.
The Power of Snekmer
Several research studies have already demonstrated the efficacy of Snekmer in protein sequence fingerprinting. For instance, Snekmer was used in a supervised mode to automatically build and evaluate models for nitrogen-cycling protein families. Different k-mer lengths and alphabets were tested, and the models performed well overall, with some variations in performance for individual families. In the second scenario, Snekmer was used in an unsupervised mode to cluster protein sequences based on their kmer profile similarity. The resulting clusters show good agreement with known functional families and can be visualized as a network. Additionally, the performance of Snekmer is compared with the MMSeqs2 clustering method on a larger dataset, and Snekmer shows reasonably good performance for annotation purposes, which is slightly lower than MMSeqs2, but Snekmer can perform reasonably well for annotation purposes.
Future Directions and Potential Applications
Snekmer opens up new avenues for protein sequence analysis and has the potential to impact various fields, including drug discovery, protein engineering, and evolutionary biology. Researchers can explore novel uses for modified microorganisms by examining the biological molecules found in microbes. The seamless integration of Snekmer into high-performance computing environments simplifies its installation process. Additionally, the integration of a novel application into the DOE KBase architecture allows users to annotate their genome and metagenome sequences. This integration enhances researchers’ ability to accurately simulate the effects of engineered bacteria, encompassing both environmental impacts and their potential benefits for crop health and bioproduction. Snekmer further aids researchers in analyzing patterns observed in microbiomes and the evolutionary processes of bacteria.
Conclusion
The inability to predict the function of a significant portion of bacterial protein sequences using current methods poses a significant obstacle in our understanding of intricate systems such as soil microbiomes. The prevailing protocols heavily rely on pair-wise alignments, which become not only computationally challenging as databases expand but also present difficulties in analysis. The effectiveness and precision of alignment-based models for protein families are contingent upon the training sets used initially, which in turn may become outdated as new sequence variations are discovered. Consequently, numerous bacterial proteins remain functionally uncharacterized or are simply assigned generic functions based solely on taxonomic information.
Snekmer represents a significant advancement in protein sequence fingerprinting, offering a scalable and accurate solution by leveraging amino acid recoding techniques. With its enhanced pattern recognition capabilities and flexible workflow, Snekmer empowers researchers to delve deeper into protein structure, function, and evolution.
Article Source: Reference Paper | Reference Article | Snekmer is Available at GitHub
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.