Following a single antibiotic course, the researchers discover how gut commensals acquire antimicrobial resistance genes (ARGs). According to the researchers, a single round of antibiotics may be enough to drive ARG acquisition and occurrence in gut commensals throughout the duration of a mammalian lifetime.
Antibiotics are frequently used to treat infections and ensure surgical treatments are safe. However, their widespread usage has resulted in the rise and spread of antibiotic-resistant bacteria, culminating in an “arms race” in whichever more powerful medications are required.
The effect of antibiotic treatment on the microbial community inhabiting the gut of mice was investigated by researchers from the Systems Ecology research group at the Luxembourg Center for Systems Biomedicine (LCSB) and the Department of Life Sciences (DLSM), as well as the Molecular Disease Mechanisms group at DLSM.
Their research, published in Nature Communications, shows that some bacteria are more likely than others to acquire antibiotic resistance genes. The researchers also go over the fundamental mechanisms of the gut microbiome’s short-term evolution of antibiotic resistance.
Analyzing the Gut Resistome in a Mouse Model
Antibiotic use, both in human disease treatment and animal husbandry, has spurred the global spread of antimicrobial resistance (AMR). Many bacteria have evolved resistance to several antibiotic classes, making it impossible to cure infections completely, which leads to the rise in the number of deaths globally. According to a recent study published in The Lancet, the worldwide burden of drug-resistant infections was expected to be around 5 million deaths in 2019, with AMR being the primary cause of about 1.3 million deaths.
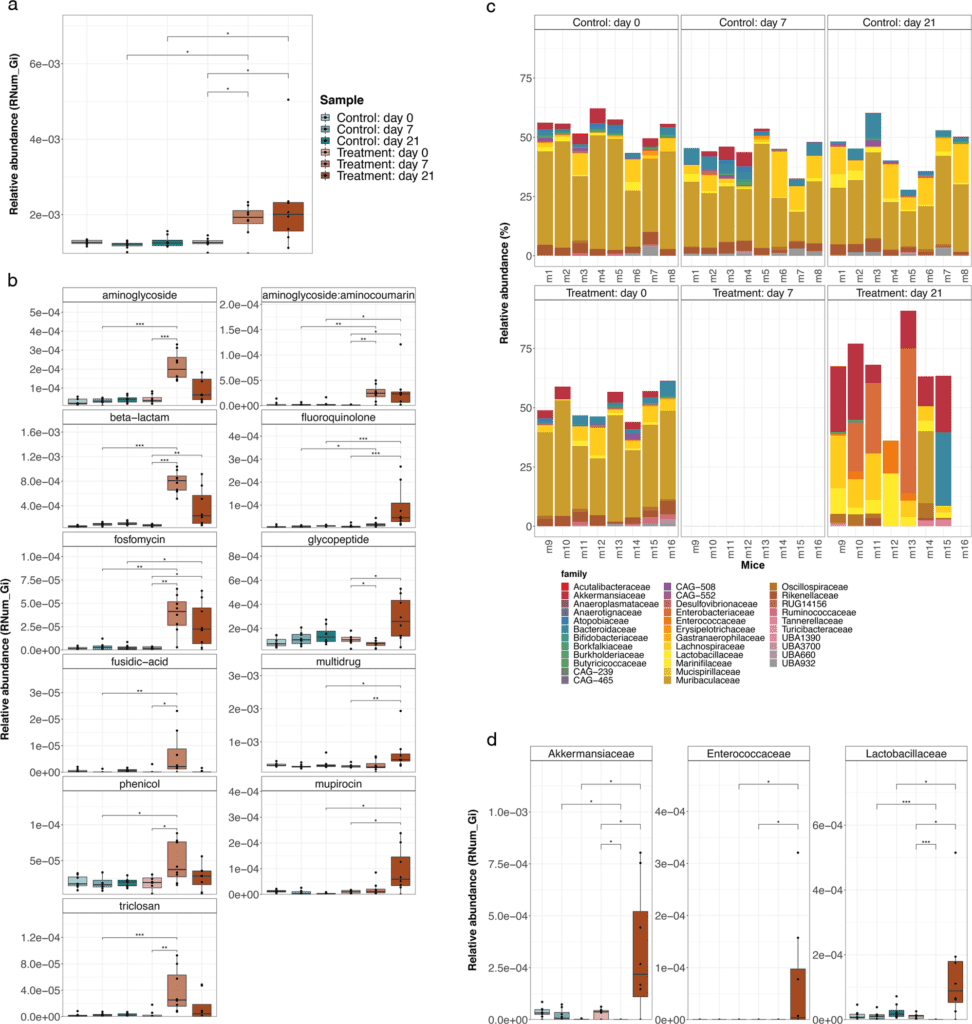
Image Source: Evolution of the murine gut resistome following broad-spectrum antibiotic treatment
The current trend is clearly not downwards, with COVID-19 further fueling the issue of antimicrobial resistance, leaving us on the course of 10 million deaths per year by 2050. This is why AMR is currently referred to as the ‘silent pandemic.’ However, there is still a lot to learn about its evolution, timescales and transmission.
Prof. Paul Wilmes, Head of the Systems Ecology Group
Researchers from the University of Luxembourg developed a mouse model to understand antibiotic resistance progression mechanisms better. They explored how some bacteria obtain antimicrobial resistance genes by treating a group of mice with a broad-spectrum antibiotic cocktail—representative of preoperative procedures—and tracking the effect on their gut flora over time.
We designed our study to understand at which stage the resistance genes are acquired and how resistant pathogens can emerge after a single antibiotic course.
Dr. Laura de Nies, post-doctoral researcher in the Systems Ecology group and co-first author of the publication
Antibiotic-Induced Gut Microbiome Makeup
The gut microbiome of antibiotic-treated mice changed dramatically, according to the researchers. While the treatment reduced the number of resident bacteria, Akkermansia muciniphila and members of the Enterobacteriaceae, Enterococcaceae, and Lactobacillaceae groups remained resistant to antibiotics.
Dr. Elisabeth Letellier, head of the Molecular Disease Mechanisms group
Interestingly, we already know that these bacteria are enriched in the gut of Parkinson’s disease patients and are associated with other chronic diseases. The fact that they are more resistant to antibiotics shows how widespread the implication of AMR can be and how important it is to better understand the underlying mechanisms.
Antimicrobial Resistance Genes Pile-up
Aside from alterations in gut microbiome composition, the researchers discovered that antibiotic-treated mice had much higher abundances of antimicrobial resistance genes. More precisely, they observed an increase in genes conferring resistance to three of the four antibiotics delivered, suggesting that these genes were acquired through time rather than being encoded within the bacteria DNA.
Our results show that the selective pressure of the administered antibiotics may lead to real-time evolution of antimicrobial resistance within the gut microbiome. It means a single treatment may already be enough to drive change within the microbial community and to lead to the acquisition of new resistance genes by some of the bacteria.
Dr. Susheel Bhanu Busi, member of the Systems Ecology group and co-first author of the publication.
Mobile Genetic Elements (MGE) Spreads Antimicrobial Resistance
Bacteria can acquire antimicrobial resistance through two genetic mechanisms: spontaneous mutations or the accumulation and transmission of resistance genes via mobile genetic elements (MGEs). These elements are genetic materials that can be passed from one species to the next. They encourage horizontal gene transfer, which is the transmission of resistance genes between bacterial populations.
The involvement of MGEs was studied to better understand the mechanism leading to the reported increase in antimicrobial resistance genes in antibiotic-treated mice. Integrons, a mobile genetic element, was discovered to play a critical role in mediating antibiotic resistance to the antibiotic cocktail provided.
The resistance genes found in our antibiotic-treated mice were mostly conferred by integrons, highlighting an underrated genetic mechanism for the transmission of antibiotic resistance. These results shed light on the short-term processes shaping the composition of antibiotic-exposed communities and the evolution of antimicrobial resistance.
Prof. Paul Wilmes
Because many of these integrons were found in genomes from antibiotic-resistant bacterial families, including Akkermansiaceae and Enterobacteriaceae, the findings highlight the relevance of certain taxonomic groups.
The study emphasizes the key role of specific bacteria and, given the association of these bacteria with some chronic diseases, we need to keep exploring the role of integrons in facilitating antimicrobial resistance within and beyond this taxon.
Prof. Paul Wilmes
Story Source: de Nies, L., Busi, S.B., Tsenkova, M. et al. Evolution of the murine gut resistome following broad-spectrum antibiotic treatment. Nat Commun 13, 2296 (2022). https://doi.org/10.1038/s41467-022-29919-9 https://wwwen.uni.lu/university/news/latest_news/antibiotics_impact_gut_microbiome_and_antimicrobial_resistance
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.