The researchers from the University of Central Florida identified promising drug candidates using an AI-based tool, “AttentionSiteDTI,” that uses natural language to model drug-target protein interactions.
Drug discovery has become a major bottleneck in modern medicine. Researchers have developed several methods to speed up the process in recent years, such as high-throughput screening (HTS). HTS involves testing thousands of chemicals against a specific biological target.
However, HTS is expensive, time-consuming, and often yields false positives. This shows that only a subset of the compounds tested will yield positive results.
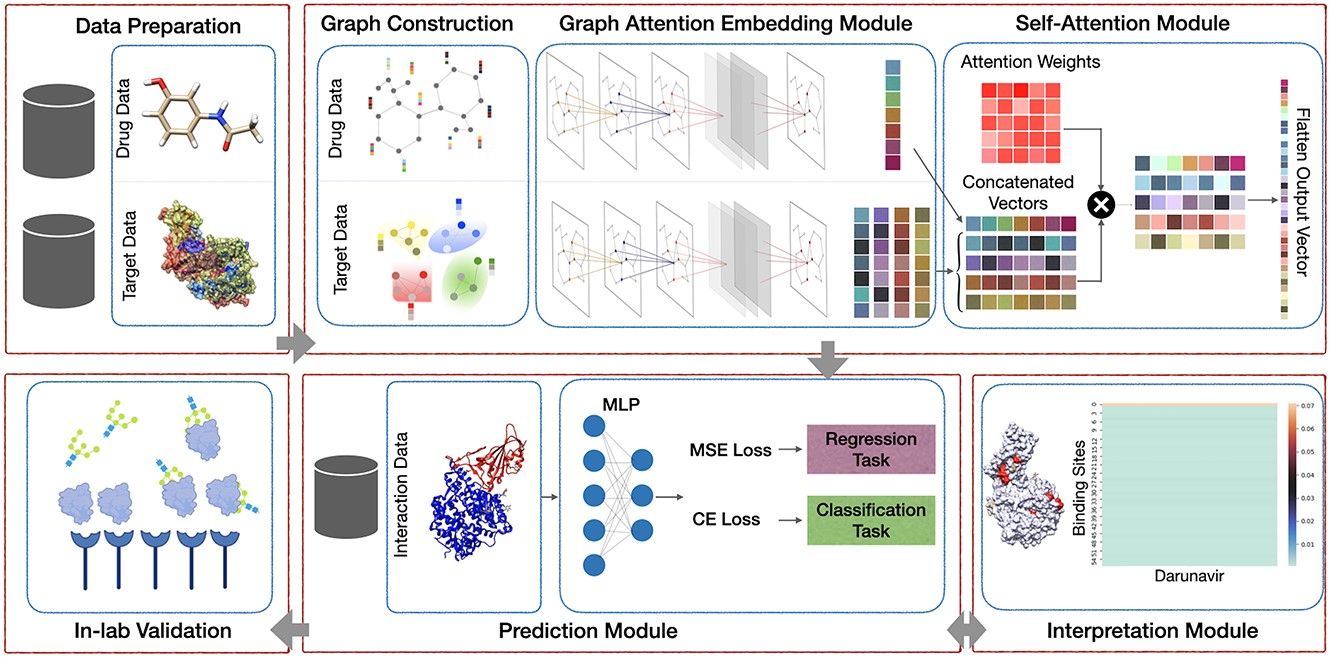
A new artificial intelligence-based drug screening process developed by the University of Central Florida researchers aims to accelerate the development of life-saving drugs. A natural language modeling approach that models drug-protein interactions led to a 97% accuracy rate in identifying drug candidates with high potential.
Drug-protein interactions are represented by words for each protein binding site, and deep learning is used to extract the features that govern their complex interactions.
According to the study’s co-author Ozlem Garibay, an assistant professor in UCF’s Department of Industrial Engineering and Management Systems, since AI is becoming more accessible, it can now be used for this purpose. There are so many variations of proteins and drug interactions that can be tried out, and it can be determined which is more likely to bind.
Drug Target Interaction Prediction (DTIP)
Drug target interactions have been widely studied in recent years due to their importance in understanding the mechanism of action of drugs. DTIP aims to predict potential drug targets based on known drug-protein interactions.
In this work, the researchers propose a novel method called AttentionSiteDTI to predict drug-target interactions. The researchers first construct a heterogeneous network consisting of protein-drug pairs and then apply attention mechanisms to learn the relationship between proteins and drugs.
The model developed by the researchers, AttentionSiteDTI, is the first of its kind that can be interpreted using the language of protein binding sites.
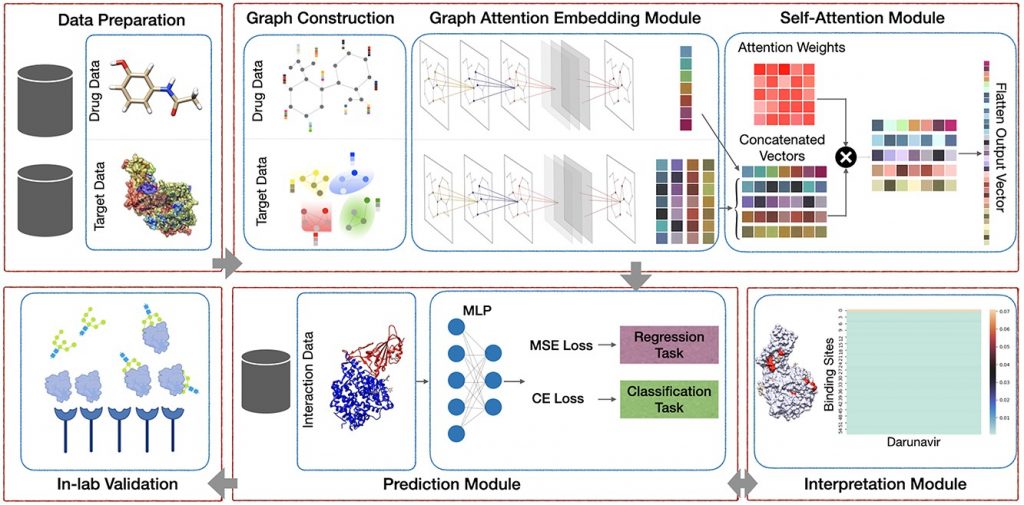
Image Source: https://doi.org/10.1093/bib/bbac272
A key element of determining a drug’s effectiveness is identifying key protein binding sites and their functional properties. This study will help identify those binding sites.
By devising a self-attention mechanism, the researchers were able to make the model learn what parts of the protein interact with drug compounds and achieve state-of-the-art prediction accuracy.
The self-attention ability of the model works by selectively focusing on the most relevant parts of the protein.
To validate their model, researchers measured binding interactions between compounds and proteins in the lab and then compared the results with their computational predictions. Since drugs to treat COVID are still in demand, the experiments also tested and validated compounds that bind to a spike protein in SARS-CoV2.
According to Garibay, the high degree of agreement between lab and computational predictions underscores the potential of AttentionSiteDTI for pre-screening and repurposing potential drugs.
In the view of Sudipta Seal, the study’s co-author and chair of UCF’s Department of Materials Science and Engineering, the high-impact research to address COVID-related discovery was only possible because of the interdisciplinary collaboration among materials engineering and AI/ML and Computer Scientists.
According to Dr. Mehdi Yazdani-Jahromi, lead author of the study and a doctoral student at the University of Central Florida’s College of Engineering and Computer Science, the work is the first step in paving a new direction for drug pre-screening.
Final thoughts
With AI, researchers can identify drugs more accurately to respond faster to new diseases. Using this method, researchers can also pinpoint which site of a virus’s protein is the most effective for designing drugs.
Further, the researchers are looking forward to using the power of artificial intelligence to design novel drugs. In order to be prepared for a pandemic, this is a logical next step.
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.