Scientists from the Broad Institute have made a groundbreaking discovery in the field of neuroscience by mapping the locations of hundreds to thousands of cell types across a mammalian brain. Two research teams at the Broad Institute made this discovery by using spatial transcriptomics to create detailed atlases of the mouse nervous system, paving the way for similar efforts in humans. The study is part of a larger effort to map all the brain’s cell types, which offers hope of one-day tracing brain activity to specific cells and understanding how the brain works.
Thousands of specialized cell types form the building blocks of the brain
The mammalian brain contains thousands of highly specialized cell types. These cells have differences in their molecular biology, structure, and function. They combine to form hundreds of distinct brain areas with particular roles. Advances in technology have recently allowed the mapping of cell types in individual brain regions. New techniques have also begun uncovering how these cell types organize spatially across brain structures. However, a complete inventory linking all specialized brain cell types to the exact structures they occupy does not yet exist. Ongoing research continues to uncover more about the vast diversity of cell types that make up the complex mammalian brain.
The Technological Marvels Behind the Mapping
The collaborative efforts of the Macosko and Chen labs were instrumental in devising a spatial transcriptomic approach meticulously implemented across the entirety of the mouse brain. Expertise from both the Broad’s Genomics Platform and the Hail support team, contributors to the open-source tool for scalable genomic analysis, played a pivotal role in the successful execution of this groundbreaking study.
The scientists pinpointed numerous distinct cell populations, meticulously plotting their positions throughout the entire tissue with nearly cellular-level precision. Their calculations suggested that approximately 90 percent of the mouse brain’s cell types were encompassed in their analysis, revealing a rich diversity within often-overlooked regions of the brain. The researchers developed an online browser to host and disseminate their datasets to facilitate collaboration and knowledge sharing within the scientific community.
The team analyzed the complete transcriptome of cells from nearly 100 regions across the mouse brain using high-throughput single-nucleus RNA sequencing, the preferred approach for efforts to create a human brain atlas. This resulted in more than four million profiles of gene activity, which they clustered into nearly 5,000 unique cell populations, most of which were neuronal cells.
The team subsequently employed a technique pioneered in the Chen and Macosko labs, namely Slide-seq. This method involved transferring 101 consecutive sections, spanning the entirety of a single mouse brain volume, onto arrays of beads coated with distinct DNA barcodes. These barcodes selectively adhered to the mRNA transcripts within the brain tissue. Subsequently, the team sequenced these transcripts and aligned the spatial data with an established 3D reference atlas. This strategic alignment allowed them to attribute each transcript to a specific brain structure, resulting in the mapping of over 1.7 million cells.
To create a comprehensive atlas of the entire mouse brain, the researchers amalgamated detailed and well-sampled cell-type profiles obtained from the single-nucleus sequencing dataset. This approach enabled them to locate each cell type in every section precisely, culminating in a meticulous and exhaustive representation of the mouse brain’s intricacies.
Furthermore, the dataset underwent thorough analysis, leading to the identification of two or three marker genes per cell population. These marker genes offer a unique means of distinguishing nearly all the distinct cell populations within the brain.
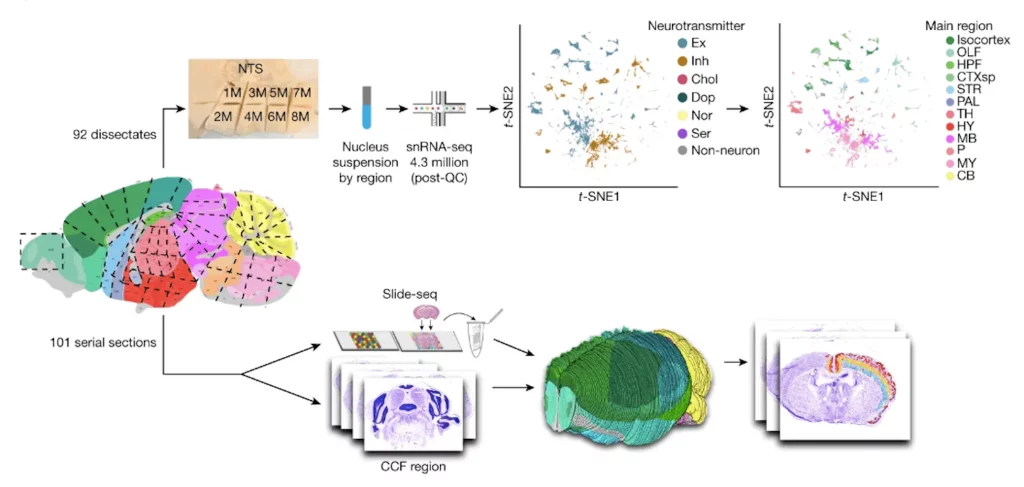
Image Source: https://doi.org/10.1038/s41586-023-06818-7
Implications for Neuroscience and Beyond
Accelerating Discoveries in Neurological Disorders
This pioneering research opens new doors for understanding and treating neurological conditions. The findings enable more targeted medical therapies by illuminating specific cell types linked to different disorders. Beyond furthering comprehension, this work may also lead to precision treatment breakthroughs for various neurologic diseases.
Navigating Neurodevelopmental Pathways
Comprehending the diverse ensemble of cell types in the mammalian brain provides a roadmap for unraveling neurodevelopmental trajectories. The research highlights the intricate cellular changes underlying brain maturation. These insights shed light on cognitive growth and conditions impacting the burgeoning brain.
Conclusion
This impactful study makes strides in demystifying the brain’s complexity. The exhaustive cataloging of cell types constructs a detailed framework of the mouse nervous system, clearing the path for comparable human cartography. By enhancing the capacity to trace brain activity to discrete cells, the research constitutes a watershed moment for understanding this organ’s inner workings. The cellular atlas of the brain promises hope of illuminating both neurological disorders and development.
Story Source: Reference Paper | Reference Article
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.