Recent breakthroughs in AI-based protein structure prediction have enormous potential to accelerate discoveries across the life sciences. However, major barriers persist in making these advanced tools accessible to non-experts without specialized computing resources. To address this gap, researchers from Joint BioEnergy Institute, US, have developed Foldy – an intuitive web application that puts powerful structural biology capabilities into any biologist’s hands. This article explores Foldy’s origins, key features, and example use cases highlighting its transformative possibilities.
The Promise and Challenges of AI-Driven Structure Prediction
In just a few years, AI models like AlphaFold have achieved dramatic leaps in accurately predicting protein structure from sequence. These methods hold tremendous promise for rational design, functional annotations, and revealing new biology.
However, major obstacles remain in applying them. Predicting structures requires heavy computing power – both GPUs and storage for the massive neural networks. This places AI-based structure prediction out of reach for many biologists.
Making such revolutionary capabilities truly accessible to non-experts is essential for catalyzing discoveries. How can we put these tools into any researcher’s hands without specialized expertise or resources?
Introducing Foldy – Democratizing Structural Biology
To overcome these barriers, researchers built Foldy, an easy-to-use web application for key structural biology workflows:
- Protein structure prediction with AlphaFold
- Domain annotations with Pfam
- Ligand docking with AutoDock Vina and DiffDock
Foldy provides an intuitive interface and multiple lightweight deployment options accessible even to non-experts. This empowers researchers to harness advanced AI tools on demand without investing in computing infrastructure.
Key Capabilities and Interface Design
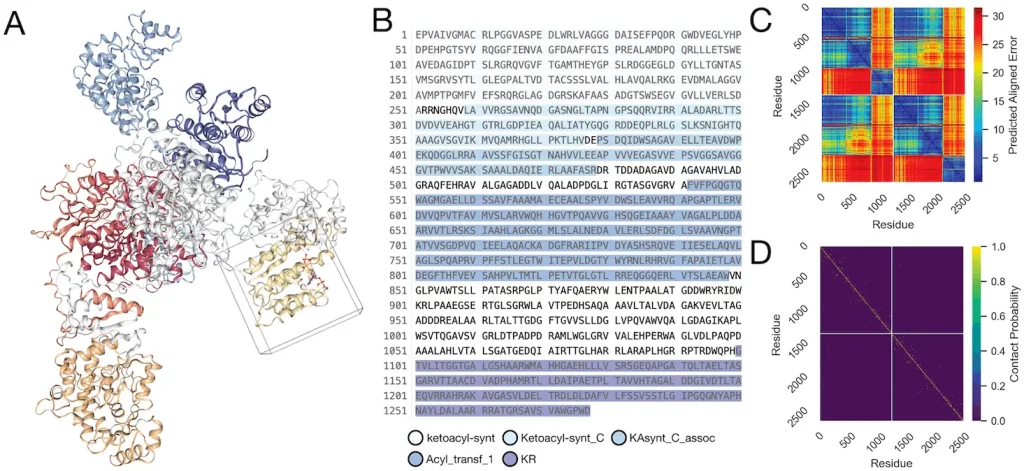
Foldy enables users to easily predict structures for proteins up to 6000 amino acids with AlphaFold Multimer. Researchers can visualize structures alongside Pfam domain annotations.
Integrated small molecule docking with AutoDock Vina and DiffDock allows screening ligand interactions. The Contacts view reveals predicted inter-protein contacts and interface residues to model complexes.
Foldy provides organization and sharing capabilities for managing prediction projects. Users can build collaborator teams, categorize entries, export structures, and make projects public for open data.
The responsive web interface allows use on any device. Slick visualizations, drag-and-drop uploads, and intuitive workflows lower adoption barriers for non-experts.
Deployment Options for Flexible Accessibility
A key focus of Foldy is providing multiple options for deploying the application:
Managed Cloud: Get running in minutes on Google Cloud through Kubernetes without any infrastructure management.
Custom Cloud: Deploy Foldy on cloud platforms like AWS or GCP for more control.
Hybrid: Utilize local HPC/GPU resources alongside cloud-based processing for custom needs.
Local: For large institutions, run entirely on-premises for full customization and privacy.
These flexible options make Foldy accessible to users at any scale and adaptable to specific needs.
Case Studies Demonstrating Scientific Utility
Foldy has already enabled a range of research applications, highlighting its capabilities:
Enzyme Engineering: Researchers used Foldy to iteratively predict mutant structures and dock ligands to redesign an enzyme’s substrate specificity.
Complex Structure Predictions: Scientists predicted megasynthase complexes over 1000 AAs to evaluate chimeric protein function based on predicted contacts.
Functional Annotations: By co-folding unknown proteins with predicted interactors, researchers derived structural insights into protein localization and function.
The integrated workflows and organizational features accelerated these projects.
Comparisons to Existing Structural Biology Tools
Foldy combines AlphaFold predictions with downstream analysis lacking in other open-source tools. Closed-source web tools offer similar capabilities but restrict access behind paywalls or limited trials.
Among open-source options, Foldy provides the most end-to-end features while maximizing accessibility via its interface and deployment options. These advantages make Foldy a practical daily driver for structural biology.
Conclusion
By lowering barriers to state-of-the-art structure prediction, Foldy promises to vastly expand access to transformative AI capabilities. Its intuitive design and flexible deployment empower any researcher to harness these tools for accelerated discovery.
Ongoing enhancements to further improve usability will help drive adoption. But already, Foldy represents a critical step towards democratizing structural biology through user-friendly software.
Story source: Reference Paper | Foldy can be accessed at Website | GitHub
Follow Us!
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.