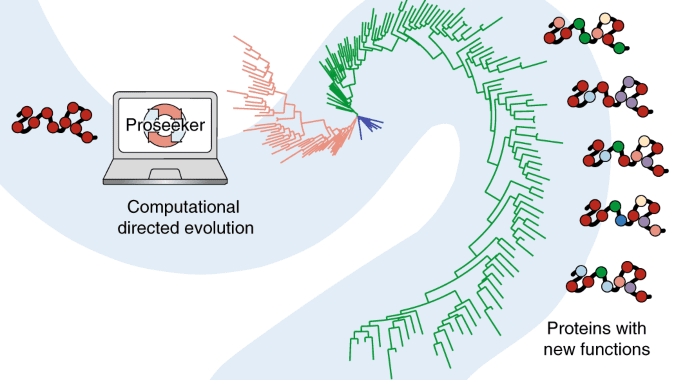
The researchers from Curtin University, Australia, and the University of Western Australia developed a system to carry out the complete process of directed evolution in silico. Thanks to revolutionary software that mimics evolution, the building blocks of life-saving treatments might be produced in days rather than years.
According to the research published this week in Nature Chemical Biology, a novel computational program ‘Proseeker’ replicates natural selection processes to produce proteins with a variety of pharmaceutical and home applications.
The enzymes present in the laundry detergent, the insulin in diabetic prescription, and the antibodies used in cancer therapy are all generated in the lab via a time-consuming process known as directed evolution.
Laboratory evolution imitates natural evolution by creating mutations in naturally-sourced proteins and choosing the finest mutants, which are then modified and chosen again in a time-consuming and tedious process that results in usable proteins.
The ARC Centre of Excellence in Synthetic Biology has now developed a technique to use a computer to undertake the entire process of guided evolution. The researchers applied iterative computational cycles of mutation and evaluation to predict mutations that confer high-affinity binding activities for DNA and RNA to an initial de novo designed protein with no inherent function. Beneficial mutations revealed nucleic acid recognition modes not previously seen in natural proteins, demonstrating how computational guided evolution might unlock novel molecular capabilities.
Furthermore, the mechanism by which new functionalities were acquired is quite similar to natural evolution. It can give insight into the effects of mutation rate, population size, and selective pressure on macromolecule functionalization in nature. This in silico method can cut the time it takes from months or even years to just a few days.
To demonstrate how valuable this method may be, the researchers provided a protein with no function, the capacity to bind DNA.
“Proteins that bind DNA are currently revolutionizing the field of gene therapy where scientists are using them to reverse disease-causing mutations,” says Professor Rackham, the leading author of the research. "So this could be of great use in the future."
Story and Background Image Source: Raven, S.A., Payne, B., Bruce, M. et al. In silico evolution of nucleic acid-binding proteins from a nonfunctional scaffold. Nat Chem Biol (2022). https://doi.org/10.1038/s41589-022-00967-y https://news.curtin.edu.au/media-releases/rebooting-evolution/
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.