Researchers from Göttingen University have found a protein switch that controls enzymes in pathogens. An allosteric lysine–cysteine redox switch with a covalent NOS bridge modulates enzyme activity in response to redox changes.
Proteins are the “workhorses” of all living cells and the chemical foundation of life. These little machines are required for performing various activities in the organism, such as converting food into energy, managing movement, brain activities, and fighting off bacteria and viruses. Protein function is closely regulated in response to changing situations, like when an organism is stressed and has to adapt. Researchers from the University of Göttingen recently found a brand-new family of proteins that work as a switch to control biological activity. They’ve now evaluated all known protein structures and discovered that this group can be found in all domains of life, including viruses, bacteria, fungus, plants, and mammals, including humans. The impact of the study is far-reaching for the treatment of a variety of disorders. The research findings were published in the journal ‘Nature Chemical Biology.’
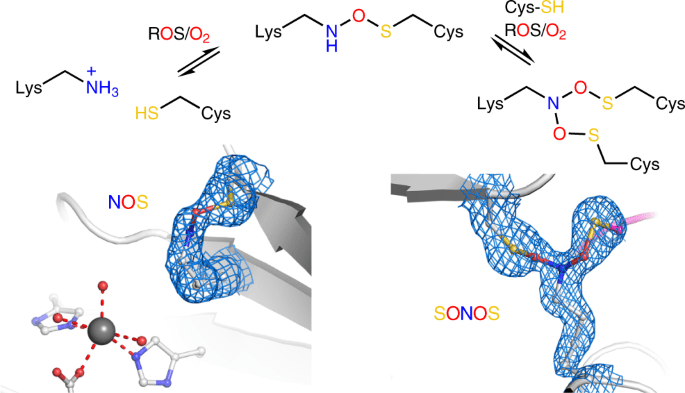
The novel protein switch was first identified in a protein from the human infection Neisseria gonorrhoeae and is called a “NOS switch” because it connects Nitrogen, Oxygen, and Sulphur atoms. The issue, however, remained as to whether or not this switch was common in nature. The researchers have now combed through a publicly accessible database of known protein structures, searching for NOS switches that had previously gone unnoticed. Dr. Fabian Rabe von Pappenheim of the University of Göttingen performed the computer study, which yielded hundreds of hits, each of which was carefully examined. “Investigating these structures was an exciting endeavor. It was like traveling into the unknown for us,” says Professor Kai Tittmann, University of Göttingen, the lead author of the study.
The researchers systematically mine the available protein structure database for proteins with undetected lysine–cysteine cross-links, discovering previously unidentified NOS bridges in proteins from all domains of life.
Image Source: Widespread occurrence of covalent lysine–cysteine redox switches in proteins.
The new NOS switch has been discovered in all domains of life and is frequently present in the regions of proteins that are required for biological activity. Surprisingly, this switch is found in a number of proteins from some of the most lethal human viruses, including a critical SARS-CoV-2 coronavirus enzyme. In fact, the recently approved antiviral treatment for individuals with mild to moderate Covid-19 who are at high risk of developing severe illness, targets this switch. Furthermore, the researchers found numerous novel chemical versions of the NOS switch, which is a widely utilized regulatory platform in biology. The discovered proteins are involved in practically every aspect of cellular activity, including gene expression, cell-to-cell signaling, and metabolism.
Reactive oxygen species (ROS) play a crucial role in redox signaling in all aspects of life, regulating cell growth, development, metabolism, aging, and the response to stress, such as pathogen infection. ROS, when elevated, causes oxidative stress, which has been linked to a variety of illnesses, including cancer, neurological diseases, inflammation, and autoimmune disorders.
Redox switches such as NOS and SONOS are ubiquitous regulatory elements in proteins that reversibly modify protein activity in response to changing redox circumstances, such as oxidative stress. The involvement of lysines in enzyme catalysis and/or binding of enzymatic substrates, nucleic acids, and effectors increases organisms’ chemical repertoire to deal with changing redox circumstances and establishes a new universal biological regulating principle. Aside from novel medicinal uses, the discovered design principles of naturally formed NOS/SONOS switches are expected to influence peptide and protein design; in particular, the recently discovered branching SONOS cross-link has a lot of promise. The occurrence of sulfenamide molecules as another type of lysine–cysteine redox switch should be investigated further.
Image Source: Widespread occurrence of covalent lysine–cysteine redox switches in proteins.
“We believe that the discovery of these new protein switches will be a springboard for the development of a novel class of drugs that directly targets these switches,” said Tittmann. “Many human proteins with known roles in severe diseases as well as proteins from bacteria and viruses are now known to be controlled by such switches. The newly identified switch is likely to play a central role in regulating their biological function as well.”
Story Source: Rabe von Pappenheim, F., Wensien, M., Ye, J. et al. Widespread occurrence of covalent lysine–cysteine redox switches in proteins. Nat Chem Biol (2022). https://doi.org/10.1038/s41589-021-00966-5
https://www.uni-goettingen.de/en/3240.html?id=6606
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.