The new approach, developed by Scripps Research experts, has the potential to speed up the development of numerous novel drugs.
Scripps Research scientists have developed a method for imaging where drugs hit their targets in the body across several tissues and with greater precision than ever before. The novel technique has the potential to become a standard tool in drug development.
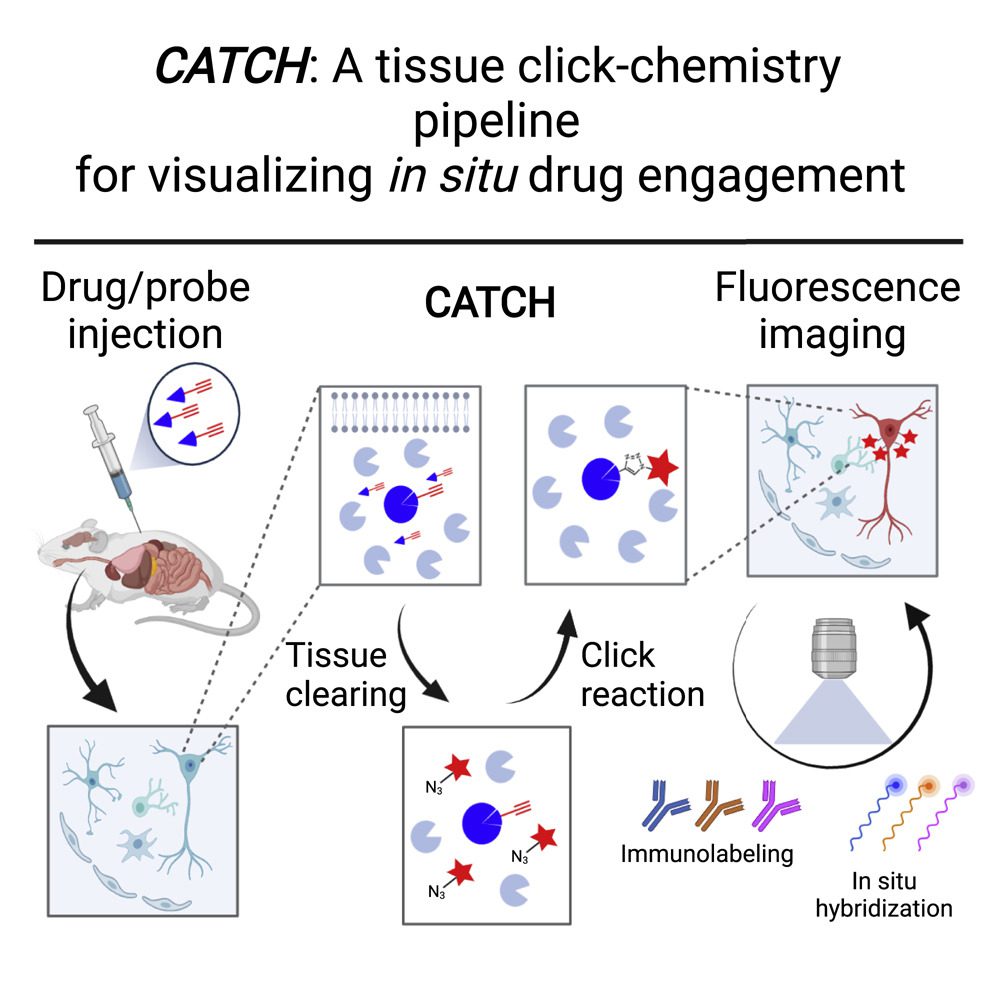
Understanding in vivo drug effects has been hampered by a lack of techniques for observing drug-target interactions at cellular resolution in intact tissue.
The new method, named CATCH ( clearing-assisted tissue click chemistry), attaches fluorescent tags to drug molecules and employs chemical procedures to increase the fluorescent signal, according to a paper published in Cell on April 27, 2022. The process was proved with numerous experimental drugs, demonstrating where the drug molecules hit their targets, even within individual cells.
According to the senior author of the study Li Ye, Ph.D., assistant professor of neuroscience at Scripps Research and The Abide-Vividion Chair in Chemistry and Chemical Biology, for the first time, the researchers were able to identify why one drug is more potent than another, or why one has a specific adverse effect while another does not.
Li Ye, PhD., believes that the development of this technique was made possible because of the unique environment at Scripps Research, where biologists and chemists routinely work together.
Understanding how drug molecules connect to their targets to produce therapeutic effects—as well as adverse effects—is a crucial element of the drug development process. On the other hand, drug-target interaction investigations have typically relied on inaccurate approaches such as bulk analysis of drug-molecule concentration in entire organs.
In the CATCH approach, small chemical handles are inserted into drug molecules. These chemical handles don’t react with anything else in the body, but they do allow fluorescent tags to be added after drug molecules have bonded to their targets. Ye and his team paired the tagging method with a technique that renders tissue relatively transparent, partly because human or animal tissue tends to disperse and obscure the light from fluorescent tags.
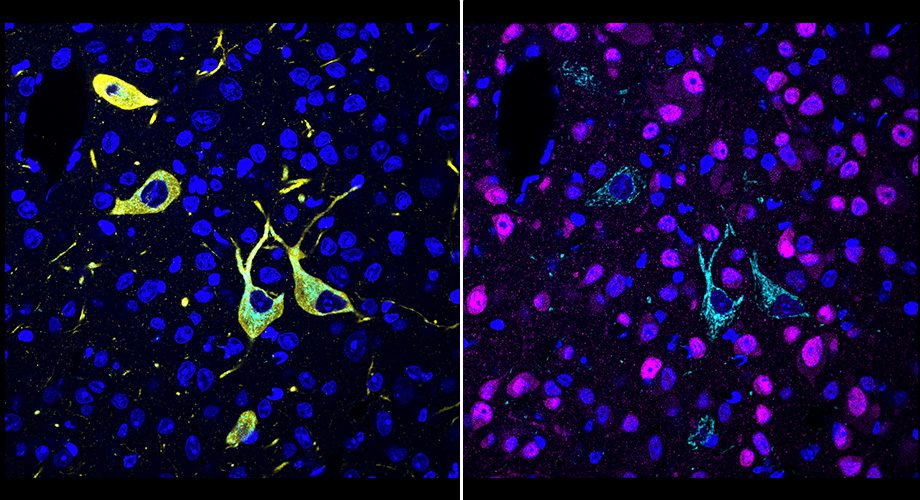
Image Source: https://www.scripps.edu/news-and-events/press-room/2022/20220427-ye-catch.html
The researchers optimized and evaluated their strategy in this initial investigation for “covalent drugs,” which bind irreversibly to their targets through stable chemical connections known as covalent bonds. Because binding is irreversible, it’s critical to make sure these medications hit their appropriate targets.
The researchers looked at a number of covalent inhibitors of a brain enzyme called fatty acid amide hydrolase (FAAH). FAAH inhibitors increase levels of cannabis compounds, notably the “bliss molecule” anandamide, and are being studied as pain and mood disorders therapies. The researchers could image where these inhibitors hit their targets in enormous volumes of mouse brain tissue at the single-cell level. They could easily discern their various patterns of target engagement.
The researchers demonstrated in one experiment that an experimental FAAH inhibitor called BIA-10-2474, which was found to cause one death and several injuries in a clinical trial in France in 2016, engages unknown targets in the midbrain of mice even when the mice lack the FAAH enzyme, pointing to the source of the inhibitor’s toxicity.
In subsequent tests, the researchers demonstrated that they could combine drug-target imaging with independent fluorescent-tagging technologies to disclose the cell types to which a drug binds, confirming the novel method’s unparalleled precision and versatility. They could also tell the difference between drug-target engagement sites in different areas of the brain. Finally, they demonstrated how slight differences in drug doses could have a big impact on the degree of target engagement in distinct brain locations.
Ye underlines that the proof-of-principle study is only the beginning. He and his colleagues want to improve CATCH so that it can be used on thicker tissue samples, possibly even whole mice. They also intend to apply the basic concept to more common non-covalently bound drugs and chemical probes.
Overall, the new technology presents a fundamental tool not only for drug development but also for basic biology.
Story Source: Pang Z, Schafroth MA, Ogasawara D, Wang Y, Nudell V, Lal NK, Yang D, Wang K, Herbst DM, Ha J, Guijas C, Blankman JL, Cravatt BF, Ye L. In situ identification of cellular drug targets in mammalian tissue. Cell. 2022 Apr 20:S0092-8674(22)00390-7. doi: 10.1016/j.cell.2022.03.040. Epub ahead of print. PMID: 35483372.
https://www.scripps.edu/news-and-events/press-room/2022/20220427-ye-catch.html
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.





