The team of researchers led by the Technical University of Munich (TUM) was able to destroy the genetic code of SARS-CoV-2 by using particular enzymes, shortly after the virus had invaded the cell. The data might be utilized to design a novel COVID-19 treatment.
The building instructions for proteins and other compounds are found in our DNA. To allow the cell to manufacture these, a transcript of these construction instructions must first be made, which may be found in the form of RNA molecules. The cells identify the transcript and put it into action.
“But there is also a mechanism that can specifically destroy this RNA and that takes place in all human cells as part of gene regulation,” explains Dr. Thomas Michler, lead author of the study. “It’s called RNA interference.”
In the process, tiny RNA fragments called siRNA are generated in the cell (small interfering RNA). These may bind to particular locations in an RNA molecule. With proteins, siRNA creates the RNA-induced silencing complex (RISC), an enzyme that cuts the target RNA.
Image Source: Targeting genomic SARS-CoV-2 RNA with siRNAs allows efficient inhibition of viral replication and spread.
The virus injects RNA into a cell
“There have long been attempts to use this mechanism to treat diseases,” explains Ulrike Protzer, head of the Institute of Virology. “There has been a lot of progress in this area in recent years. Among other things, it is now possible to stabilize the siRNA through chemical modifications in such a way that it is not broken down so quickly in the cells.”
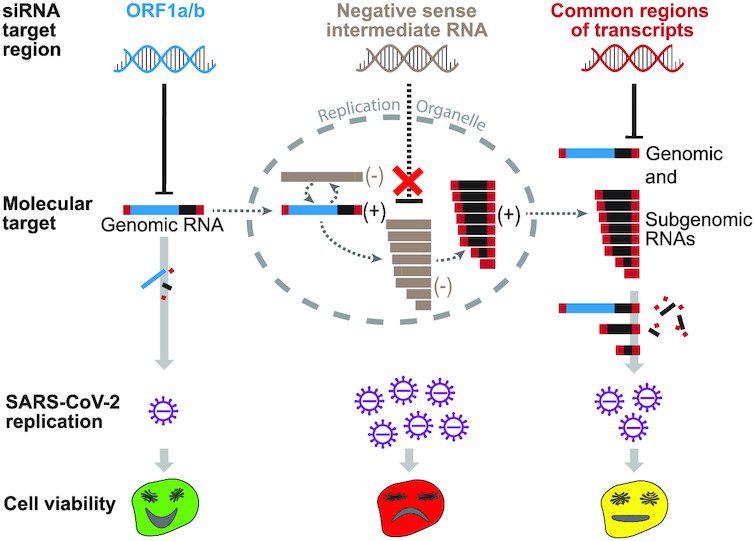
For RNA interference in SARS-CoV-2, there are two points of attack: To begin, the virus’s genome is made up of RNA, which is delivered into the infected cell and contains the genetic code for future viruses. On the other hand, subgenomic RNA molecules are generated, instructing the host cell to create viral proteins.
Beginning of the replication cycle as the most effective point of attack
Above all, the researchers needed to figure out which structures of viral RNA can be addressed the most effectively and when the therapy should occur throughout the replication cycle. “The most effective time for RNA interference is when the virus has just entered the cell,” says Shubhankar Ambike, one of the study’s primary authors. In this case, siRNAs that target the viral genome specifically outperformed siRNAs that target subgenomic RNA molecules.
The researchers also conducted studies on human lung tissue infected with SARS-CoV-2 in collaboration with colleagues from Ludwig-Maximilians-Universität and Helmholtz Zentrum München. In the experiments, they were able to corroborate their findings. The researchers are currently working on a project to establish a way to get the active chemical into the lungs as effectively as possible. The findings might potentially be used to develop treatments for other respiratory virus infections.
Story Source: Shubhankar Ambike, Cho-Chin Cheng, Martin Feuerherd, Stoyan Velkov, Domizia Baldassi, Suliman Qadir Afridi, Diana Porras-Gonzalez, Xin Wei, Philipp Hagen, Nikolaus Kneidinger, Mircea Gabriel Stoleriu, Vincent Grass, Gerald Burgstaller, Andreas Pichlmair, Olivia M Merkel, Chunkyu Ko, Thomas Michler, Targeting genomic SARS-CoV-2 RNA with siRNAs allows efficient inhibition of viral replication and spread, Nucleic Acids Research, Volume 50, Issue 1, 11 January 2022, Pages 333–349, https://doi.org/10.1093/nar/gkab1248
https://www.tum.de/die-tum/aktuelles/pressemitteilungen/details/37208
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.