Protein complex structures form the foundation for understanding the complexities of molecular biology. While deep learning-based methods have significantly enhanced the prediction of single protein structures, predicting the structures of protein complexes remains a challenge. This challenge arises from the limited availability of structural and evolutionary information for protein complexes. However, recent breakthroughs have incorporated experimental data, particularly from crosslinking mass spectrometry (MS), into deep learning models to enhance the accuracy of protein complex structure prediction.
Improving Structure Prediction with Crosslinking MS Data
Crosslinking MS is a powerful experimental technique used to identify protein-protein interactions in complex mixtures. It involves chemically crosslinking amino acids in close proximity within a protein or between different proteins in a complex. After being enzymatically digested, the crosslinked proteins are subsequently isolated from the crosslinked proteins by enzymatic digestion and mass spectrometric analysis. These crosslinking data provide valuable distance restraints that can be utilized to guide the prediction of protein complex structures.
One such deep learning model that integrates crosslinking MS data is AlphaLink. Building upon the success of AlphaFold, which revolutionized the prediction of single protein structures, AlphaLink extends the methodology to tackle protein complexes. Researchers have modified the code base and trained the network with simulated crosslinks using a soluble crosslinker called succinimidyl 4,4-azipentanoate (SDA). This modification allows the incorporation of distance restraints between specific amino acids on protein surfaces.
Impressive Performance on Challenging Targets
To assess the effectiveness of AlphaLink, it was evaluated on challenging heteromeric targets from the CASP15 competition. The results demonstrated significant improvements in prediction quality compared to AlphaFold-Multimer, even when using significantly less sampling. AlphaLink achieved similar or better results than the best-performing algorithms in CASP15. Incorporating crosslinking MS data increased the DockQ score, a measure of model quality, from 0.14 to 0.48 on average for the test set.
Notably, AlphaLink excelled in predicting complex structures involving nanobody-antigen and antibody-antigen targets. These targets often have lower co-evolutionary signals, making structure prediction more challenging. However, the experimental crosslinking of MS data played a crucial role in aiding prediction, enabling AlphaLink to generate at least medium-quality models, outperforming even the top-ranked CASP15 submissions for some targets.
Addressing Limitations and Future Directions
Although AlphaLink has shown promising results, there are still challenges to overcome. Inaccurate prediction of side chain interactions and difficulties in modeling flexible targets with few contacts in the interface are some of the limitations observed. Further adjustments can be made to address these issues, such as increasing sampling and incorporating physical terms into the loss function. Refining the fold-and-dock approach of AlphaFold and AlphaLink is also necessary to improve the prediction of specific chain interactions within protein complexes.
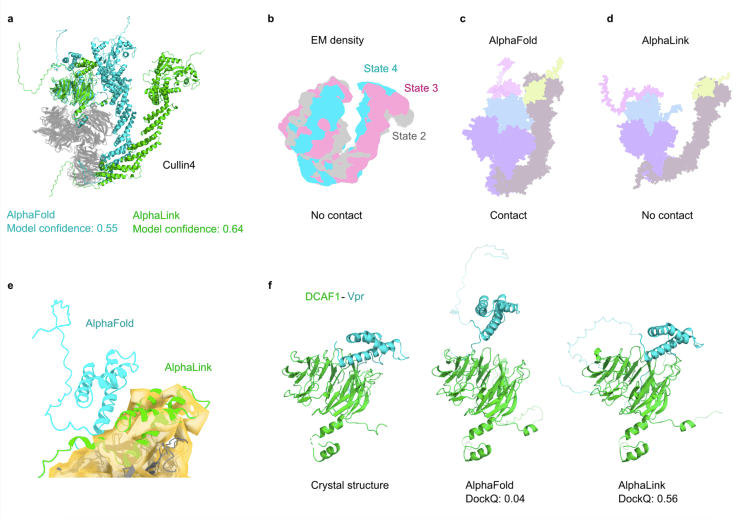
Moreover, AlphaLink’s success paves the way for broader applications in whole-cell structural investigations. By integrating crosslinking MS data from Bacillus subtilis and a viral-modified Cullin4-RING ubiquitin ligase (CRL4) complex, researchers have achieved a breakthrough in visualizing protein-protein interactions within cells at a pseudo-atomic resolution. Even a single crosslink obtained in cells drastically improves the quality of the predicted models, enabling the study of protein interactions within their native cellular context.
Modeling a Multi-Protein Complex
Further demonstrating its effectiveness, AlphaLink was applied to model the CRL4DCAF1-CtD/Vprmus/SAMHD1 complex. This protein complex plays a crucial role in controlling HIV infection by targeting viral proteins for degradation. The structural insights provided by AlphaLink revealed key interactions and allowed for the identification of important residues involved in the formation of the complex. This showcases the broad applicability of AlphaLink to different crosslinker chemistries and its potential for investigating diverse protein complexes.
Conclusion
The integration of crosslinking MS data into AlphaLink extends the capabilities of deep learning-based methods, enabling the prediction of protein complex structures. AlphaLink outperforms traditional structure prediction methods by incorporating experimental distance restraints and providing valuable insights into protein-protein interactions at a higher resolution. The impressive performance of AlphaLink on challenging targets and its potential for investigating diverse protein complexes demonstrate its importance in advancing our understanding of complex biological processes. Additionally, the application of AlphaLink in whole-cell structural investigations opens up new avenues for studying protein interactions within their native cellular context. These advancements contribute to the field of structural biology and hold great promise for guiding the development of new therapeutic strategies targeting protein complexes.
Article Source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.