The new tool “CheXzero” improves clinical AI design by overcoming the major hurdle of labeling datasets for model training.
Scientists at Harvard Medical School and Stanford University have developed an artificial intelligence diagnostic tool that can detect diseases directly from natural language descriptions of chest X-rays.
This is a major step forward in clinical AI design because most current models require laborious human annotation of enormous amounts of data before they can be fed into a model for training.
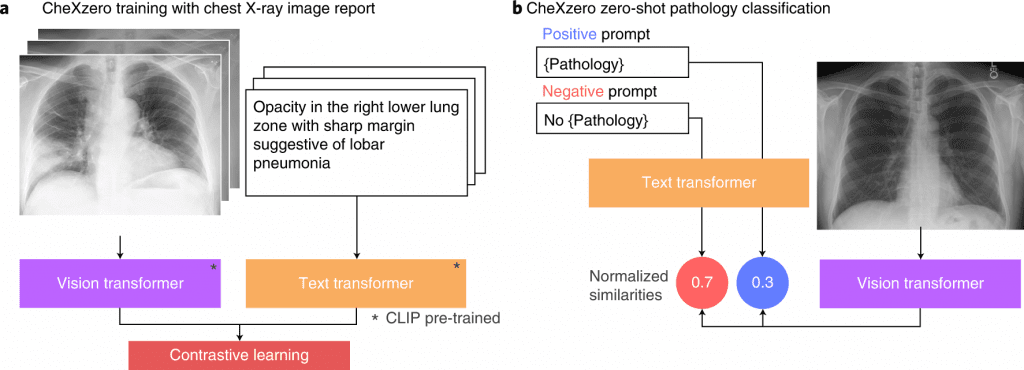
Image Source: https://doi.org/10.1038/s41551-022-00936-9
An appropriately trained machine-learning model can often outperform medical experts at interpreting medical images. However, such high performance requires that the models be trained using relevant datasets that have been meticulously annotated by experts. Based on chest X-rays without explicit annotations, the researchers show that a self-supervised model can perform pathology classification tasks as accurately as radiologists.
The model, called CheXzero, published in a study in Nature Biomedical Engineering, can detect chest X-ray pathologies on par with human radiologists.
The Hurdle of Labeled Data
AI models require labeled datasets to correctly identify pathologies when they are “trained.” It is especially challenging for medical image interpretation tasks since they require large-scale annotation by humans, which is time-consuming and expensive. The process of labeling a chest X-ray dataset, for instance, would require radiologists to review hundreds of thousands of images and provide explicit annotations to each. Recent AI models have tried to address this labeling problem by learning from unlabeled data in a “pre-training” stage, but they ultimately need labeled data to perform well.
The Solution: Unsupervised Learning
The new model, however, is self-supervised since it can learn more independently without requiring hand-labeled data prior to or after training. The model relies only on chest X-rays and English-language notes contained in X-ray reports.
According to the study’s lead investigator, Pranav Rajpurkar, assistant professor of biomedical informatics in the Blavatnik Institute at HMS, the next-generation medical AI models, which directly learn from text, are in their early stages of development. Traditionally, AI models have relied on manual annotation of thousands of images to achieve high performance. There is no need to provide such disease-specific annotations with this method.
According to Rajpurkar, with CheXzero, one can simply feed the model a chest X-ray and a corresponding radiology report, and it will learn to recognize the image and the text in the report as similar, so it will learn to match chest X-rays with their accompanying reports. A model eventually learns how concepts in the unstructured text correspond to visual patterns in images.
A dataset of more than 377,000 X-rays and 227,000 corresponding clinical notes was used to train the model. The software’s performance was assessed using two different datasets of chest X-rays and corresponding notes, one of which was collected from a foreign institution. The model was trained using a variety of datasets to ensure that it performed equally well when exposed to clinical notes that may have used different terminology to describe the same finding.
It was found that CheXzero was able to detect pathologies that were not explicitly annotated by humans during testing. Compared to other self-supervised AI tools, it outperformed them and performed with an accuracy similar to that of human radiologists.
According to the researchers, the approach could eventually be applied to imaging modalities other than X-rays, such as CT scans, MRIs, and echocardiograms.
According to study’s co-first author, Ekin Tiu, an undergraduate student at Stanford and a visiting researcher at HMS, with CheXzero, complex medical image interpretation is no longer dependent on large labeled datasets. As a driving example, the researchers use chest X-rays, but CheXzero has a capability that can be generalized to a wide range of medical settings with unstructured data, and it represents the promise of bypassing the large-scale labeling bottleneck in medical machine learning.
Benefits of CheXzero
- Allows for the design of clinical AI with minimal labeled data.
- Can save time and resources typically spent on manual annotation.
- Has the potential to improve patient outcomes by making it possible to develop AI for rare or under-studied conditions.
Final thoughts
The researchers have developed a self-supervised method using contrastive learning to detect the presence of multiple pathologies in chest X-ray images. This study shows how deep-learning models can be used to interpret medical images using large amounts of unlabeled data. This may result in a reduction of the need for labeled datasets and a decrease in clinical-workflow inefficiencies.
The team has made the model’s code publicly available for other researchers.
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.






