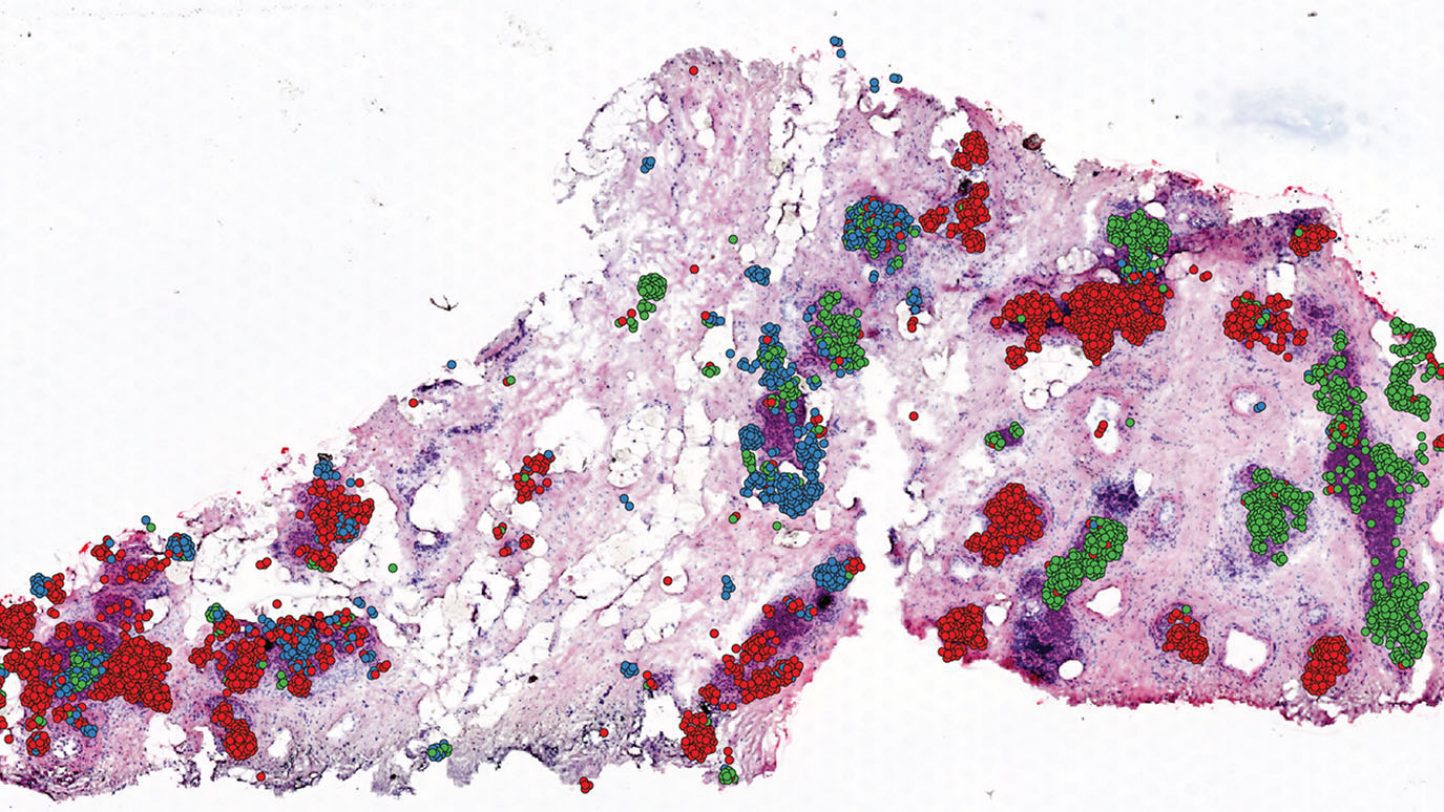
‘CellTrek’ can accurately map single cells in diverse tissue types to resolve their spatial organization. The tool identifies the location of particular cell types to drive biological insights.
Scientists at The University of Texas MD Anderson Cancer Center have created a new computational methodology that successfully combines data from simultaneous gene-expression profiling approaches to build spatial maps of a specific tissue at single-cell resolution. The generated maps can reveal new biological information about the cancer microenvironment and a variety of other tissue types.
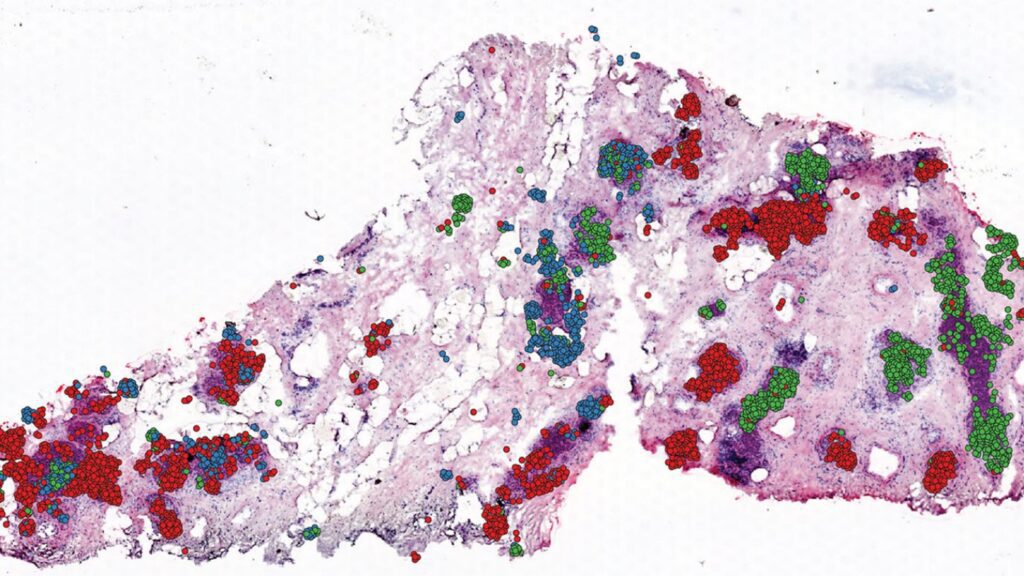
Image Source: https://www.mdanderson.org/newsroom/computational-approach-enables-spatial-mapping-of-single-cell-data-within-tissues.h00-159538167.html
CellTrek is a method that precisely pinpoints the location of particular cell types inside a tissue by combining data from single-cell RNA sequencing (scRNA-seq) with data from spatial transcriptomics (ST) tests — which quantify spatial gene expression in many small groups of cells. The scientists reported the findings of the analyses of kidney and brain tissues, as well as samples of ductal carcinoma in situ (DCIS) breast cancer.
“Single-cell RNA sequencing provides tremendous information about the cells within a tissue, but, ultimately, you want to know where these cells are distributed, particularly in tumor samples,” says Nicholas Navin, Ph.D., professor of Genetics and Bioinformatics & Computational Biology and senior author of the study. “This tool allows us to answer that question with an unbiased approach that improves upon currently available spatial mapping techniques.”
Single-cell RNA sequencing is a well-established approach for analyzing the gene expression of many individual cells in a sample, but it can’t tell you where the cells are in the tissue. ST assays, on the other hand, can detect spatial gene expression by evaluating several small groups of cells across a tissue, but they lack single-cell precision.
According to Navin, current computer methods, known as deconvolution techniques, can identify distinct cell types in ST data, but they can’t provide comprehensive information at the single-cell level.
As a result, the Navin Laboratory’s Runmin Wei, Ph.D., and Siyuan He leads the initiative to develop CellTrek as a tool that combines the unique benefits of scRNA-seq and ST tests to build accurate spatial maps of tissue samples.
The researchers revealed that CellTrek obtained the most accurate and detailed spatial resolution of the approaches assessed using publically accessible scRNA-seq and ST data from brain and kidney tissues. The CellTrek method also discerns modest gene expression changes within the same cell type, allowing researchers to learn more about the sample’s heterogeneity.
CellTrek was also used to examine DCIS breast cancer tissues in collaboration with Savitri Krishnamurthy, M.D., professor of Pathology. The scientists discovered that separate subgroups of tumor cells were evolving in unique patterns inside certain tumor sections after analyzing 6,800 single cells and 1,500 ST regions from a single DCIS sample. The ability of CellTrek to rebuild the spatial tumor-immune microenvironment within a tumor tissue was proved using a second DCIS sample.
While this method isn’t limited to examining tumor tissues, it has clear implications for a better understanding of cancer. Pathology drives cancer diagnosis, and with this tool, molecular data can be mapped on top of pathological data to allow for even more detailed tumor classifications and provide better treatment direction.
Story Source: Wei, R., He, S., Bai, S. et al. Spatial charting of single-cell transcriptomes in tissues. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01233-1
https://www.mdanderson.org/newsroom/computational-approach-enables-spatial-mapping-of-single-cell-data-within-tissues.h00-159538167.html
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.