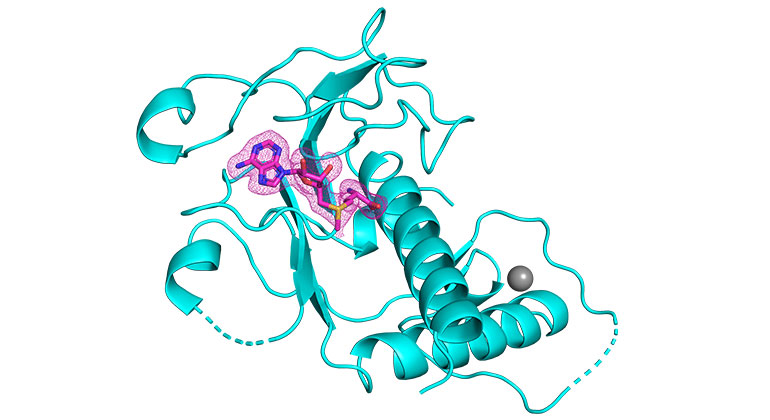
Researchers at Mount Sinai have solved the high-resolution crystal structure of a critical enzyme of SARS-CoV-2, leading to the development of new antivirals. This is a major breakthrough in the battle against SARS-CoV-2 and could lead to the development of new and more effective antiviral drugs.
Image Source: Kottur, et al; Nature Structural & Molecular Biology
SARS-CoV-2 is a deadly virus that has killed millions of people worldwide. The enzyme, known as nsp14, contains a critical region known as the RNA methyltransferase domain, which has eluded previous scientific attempts to characterize its three-dimensional crystal structure. A paper describing the novel process was published in the online edition of Nature Structural & Molecular Biology.
The research team has determined the three-dimensional structures of the SARS-CoV-2 nsp14 N7-methyltransferase core bound to three different molecules. The researchers also identify features of the methyltransferase core that are crucial for the development of antivirals and show SAH as the best scaffold for the design of antivirals against SARS-CoV-2 and other pathogenic coronaviruses.
The new structure of the methyltransferase domain of nsp14 provides insights into how to design small-molecule inhibitors that can fit into its active site and inhibit its essential chemistry. This information can be used to develop new antivirals that can be used in combination with vaccines to combat SARS-CoV-2.
Prescription antivirals that target key SARS-CoV-2 enzymes include nirmatrelvir, which targets the main protease (MPro) enzyme, and molnupiravir and remdesivir, which target the RNA polymerase (nsp12) enzyme. The development of new antivirals targeting various enzymatic activities has been accelerating in laboratories around the world, and Mount Sinai’s discovery has contributed significantly to that effort.
According to the paper’s senior author, Dr. Aneel Aggarwal, Ph.D., Professor of Pharmacological Sciences at the Icahn School of Medicine at Mount Sinai, the knowledge gained from treating HIV, such as the fact that you generally need a cocktail of inhibitors for maximum effect against the virus, is a part of what drives this research.
The Mount Sinai team has developed three crystal structures of nsp14, each with different cofactors, from which they discovered the best scaffold for antiviral design to inhibit the activity of RNA methyltransferase, which the enzyme develops and is required for the survival of the virus. The researchers believe that the antiviral would bind at the place of the natural cofactor S-adenosylmethionine, therefore preventing the chemistry methyltransferase from happening. The crystal structures developed in the study have been made public, and these will now serve as guides for biochemists and virologists around the world as they engineer these compounds.
According to Jithesh Kottur, Ph.D., a postdoctoral fellow at Icahn Mount Sinai, and a crystallographer and biochemist, the researchers were able to clear a hurdle that had prevented others in the past from creating three-dimensional crystals of the nsp14 methyltransferase domain. An approach known as fusion-assisted crystallization was employed in the study, which involves fusing the enzyme with another small protein that helps in crystalization.
According to Dr. Aggarwal, an internationally recognized structural biologist, the importance of continuing research into coronaviruses cannot be understated—because the virus evolves so quickly, it can develop resistance to the antiviral drugs that are currently available, and we must continue to develop new ones. Because nsp14 has high sequence conservation across coronaviruses and their variants (meaning it does not mutate much), the findings will help in the development of broad-spectrum antivirals for both current and future coronavirus outbreaks.
Final Thoughts
This study presents insight into the molecular basis of the SARS-CoV-2 N7 MT activity and suggests potential targets for drug discovery. Overall, the high-resolution structures of SARS-CoV-2 nsp14 N7-MTase presented here will aid in the development of new antivirals against SARS-CoV-2 and other pathogenic coronaviruses.
Story Source: Reference Paper | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.