Single-cell RNA sequencing (scRNA-seq), a rapidly developing field of RNA sequencing of single cells, has recently made a breakthrough that could revolutionize cell type annotation. Using GPT-4’s powerful language model, Columbia University Mailman School of Public Health and Duke University School of Medicine researchers have demonstrated that cell types can be accurately annotated based on marker gene information, greatly reducing the effort and expertise required for this critical step. The researchers additionally developed an R programming suite named GPTCelltype tailored for the automated annotation of cell types by GPT-4.
The Challenge of Cell Type Annotation
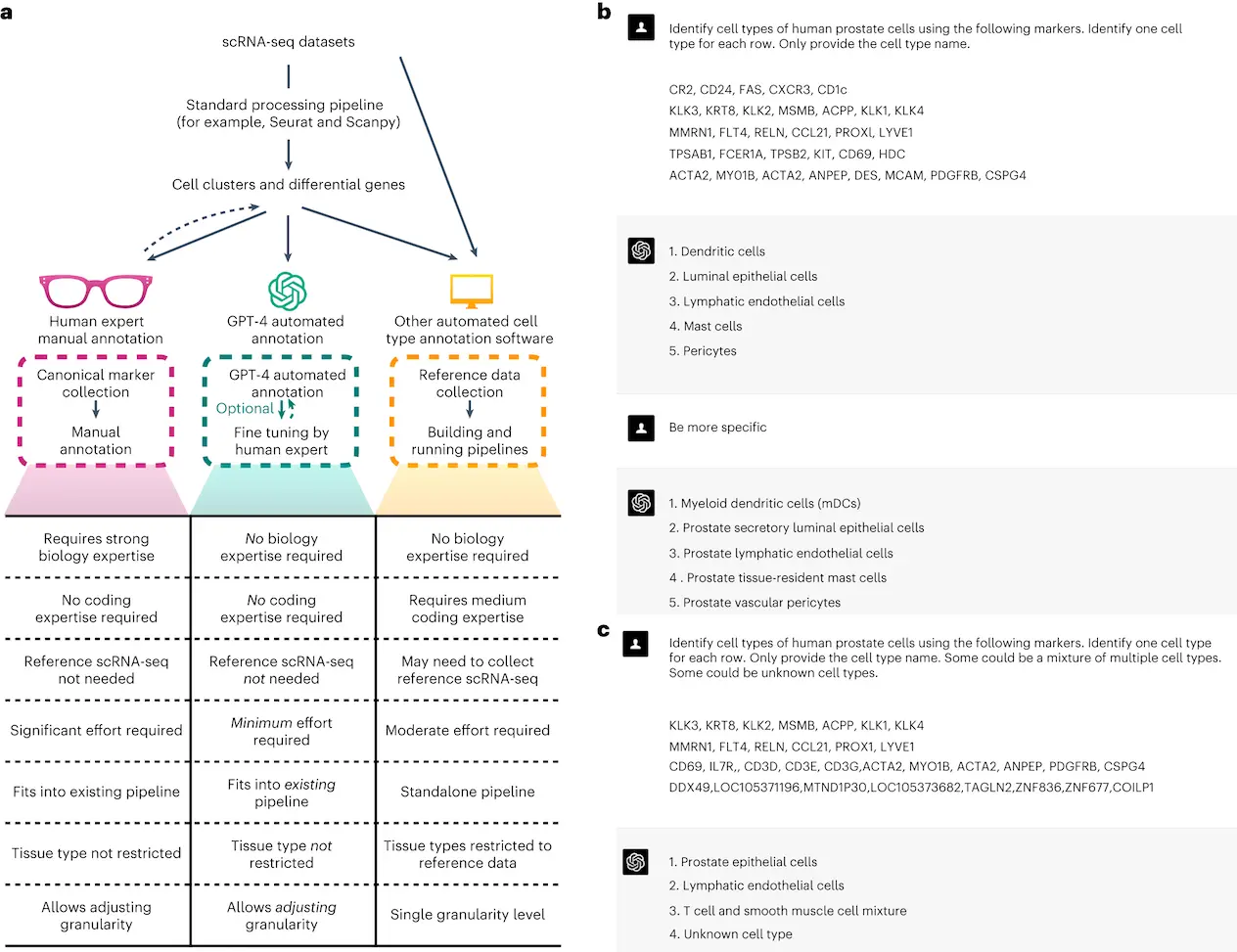
Although cell type annotation is a fundamental step in scRNA-seq analysis, it has traditionally been a long and laborious process. This analysis is typically done by a human expert by comparing highly expressed genes with canonical cell type markers. The task requires a lot of knowledge and experience.
Despite the development of automated cell type annotation methods, manual annotation based on marker genes remains widely used, raising the need for more efficient and accurate solutions.
GPT-4: A Game-Changer for Cell Type Annotation
Generative pre-trained transformers, like GPT-3.5 and GPT-4, are language models for understanding and generating languages. In recent years, they’ve shown their effectiveness in a lot of biomedical contexts, making them a good fit for scRNA-seq.
The researchers hypothesized that GPT-4 could be used to accurately annotate cell types, transitioning the annotation process from manual to semi-automated.
Evaluating GPT-4’s Performance Across Diverse Datasets
To assess GPT-4’s cell type annotation capabilities, the researchers conducted a systematic evaluation across ten datasets spanning five species and hundreds of tissue and cell types, including both normal and cancer samples.
GPT-4 was queried using GPTCelltype, a software tool developed by the researchers, which serves as an interface for the language model. The performance of GPT-4 was compared against other state-of-the-art automatic cell type annotation methods, such as GPT-3.5, CellMarker2.0, SingleR, and ScType.
As a result of the evaluation, GPT-4’s annotations were fully or partially comparable to manual annotations in more than 75% of cell types across most studies and tissues, showing its competency in generating expert-comparable annotations. In most tissues, GPT-4’s annotations and manual annotations matched well, with at least 70% full match rates.
Robustness and Reproducibility
Aside from its impressive accuracy, GPT-4 had remarkable robustness in real-world data scenarios. It could differentiate between pure and mixed cell types with 93% accuracy and known and unknown cell types with 99% accuracy.
Moreover, GPT-4 was highly reproducible, generating identical annotations for the same marker genes in 85% of cases, and showed substantial consistency with a Cohen’s correlation coefficient of 0.65 across two different versions of GPT-4.
Outperforming Existing Methods
In a head-to-head comparison, GPT-4 significantly outperformed other automatic cell type annotation methods on the basis of average agreement scores. Furthermore, GPT-4 demonstrated significantly faster performance than its competitors, largely as a result of its utilization of differential genes from standard single-cell analysis pipelines such as Seurat when using GPTCelltype as the interface.
Cost-Effective and Seamless Integration
An important advantage of GPT-4 for the annotation of cell types is its cost effectiveness and seamless integration into existing single-cell analysis pipelines. Unlike other methods, which require additional pipelines and gathering high-quality reference datasets, GPT-4 leverages its vast training data in order to enable broad applications across a wide range of tissues and cell types.
The chatbot nature of GPT-4 also allows for user-driven annotation refinement, further enhancing its versatility and accuracy.
Limitations and Recommendations
Even though GPT-4 excels at cell type annotation, there are some limitations. Because GPT-4’s training corpus isn’t disclosed, verifying the basis of annotations is hard, so human assessment is required to ensure annotation quality and reliability.
A human-mediated fine-tuning of GPT-4 may also adversely affect reproducibility due to subjectivity, and low scRNA-seq data quality and unreliable differential genes can adversely affect GPT-4’s annotation.
It is recommended that GPT-4’s cell type annotations be validated by human experts before proceeding with downstream analysis in order to reduce the risk of artificial intelligence hallucinations.
Future Possibilities: Fine-Tuning GPT-4 for Enhanced Performance
This study examined the standard version of GPT-4; however, the researchers suggest that fine-tuning GPT-4 with high-quality reference marker gene lists may enable it to improve its ability to identify cell types. Utilizing services such as ‘GPTs’ offered by OpenAI could enable this.
Conclusion
An important milestone in cell type annotation has been reached with GPT-4’s use for single-cell RNA sequencing. Researchers can use this large language model to simplify the annotation process.
This technological advancement will increase access to cell type annotation to a broader range of researchers, accelerating single-cell genomic discoveries.
Using cutting-edge language models such as GPT-4 in biomedical research and analysis holds tremendous promise for enabling breakthroughs and advances that will revolutionize many aspects.
Article source: Reference Paper | GPTCelltype open-source software package (v.1.0.0) is available with a detailed user manual in the GitHub
Follow Us!
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.