A new research reveals how a protein removes harmful chemicals from bacterial cells.
The structure of a protein that may pump hazardous chemicals out of bacterial cells has been identified by MIT researchers. Proteins like this one, discovered in E. coli, are thought to aid in developing antibiotic resistance in bacteria.
Multidrug resistance (MDR) is a severe public health problem that has the potential to sabotage years of drug research efforts and result in medication-resistant disease outbreaks. The production of transmembrane (TM) efflux pump proteins of the small multidrug resistance (SMR) family is one of the processes by which cells might become therapeutically resistant.
Unraveling the mechanism of membrane transport by EmrE requires atomic-resolution structural information for different states of the protein, as well as dynamical information on the protein and the ligands during the transport cycle to elucidate the process.
The researchers utilized nuclear magnetic resonance (NMR) spectroscopy to figure out how the structure of this protein changes as a drug-like molecule passes through it. According to Mei Hong, an MIT professor of chemistry, knowing this particular structure may make it easier to build medications that inhibit these transport proteins and enable drug-resistant bacteria to resensitize to current antibiotics.
“Knowing the structure of the drug-binding pocket of this protein, one might try to design competitors to these substrates, so that you could block the binding site and prevent the protein from removing antibiotics from the cell,” says Hong, the senior author of the study.
Image Source: High-pH structure of EmrE reveals the mechanism of proton-coupled substrate transport.
Transporters in Drug-resistance
Antibiotic-resistant bacteria can adopt a variety of techniques to avoid antibiotics, including pumping medicines out through their cell membranes. Henzler- Wildman’s team at the University of Wisconsin has been investigating EmrE, i.e., a homodimeric bacterial membrane protein that causes multidrug resistance by effluxing polyaromatic cationic substrates in a proton-coupled manner. It transports a variety of hazardous chemicals, including herbicides and antimicrobial agents, for several years.
EmrE is a member of the small multidrug resistance (SMR) transporter family of proteins. Although EmrE is not directly implicated in antibiotic resistance, additional members of the EmrE family have been discovered in drug-resistant Mycobacterium TB and Acinetobacter baumanii strains.
“The SMR transporters have high sequence conservation across key regions of the protein. EmrE is by far the best-studied member of the family, both in vitro and in vivo, which makes it an ideal model system to investigate the structure that supports SMR activity,” says Henzler-Wildman.
Image Source: High-pH structure of EmrE reveals the mechanism of proton-coupled substrate transport.
Hong’s group created a technology that allows researchers to determine the distances between fluorine probes and hydrogen atoms in proteins using NMR a few years ago. This enables the structure of a protein to be determined as it binds to a fluorine-containing molecule.
Henzler-Wildman proposed working together to explore EmrE after Hong presented a lecture about the new technology at a conference. EmrE carries a drug-like molecule, or ligand, across the phospholipid membrane, according to her research. The F4-TPP+ ligand is a tetrahedral molecule having four fluorine atoms, one at each corner.
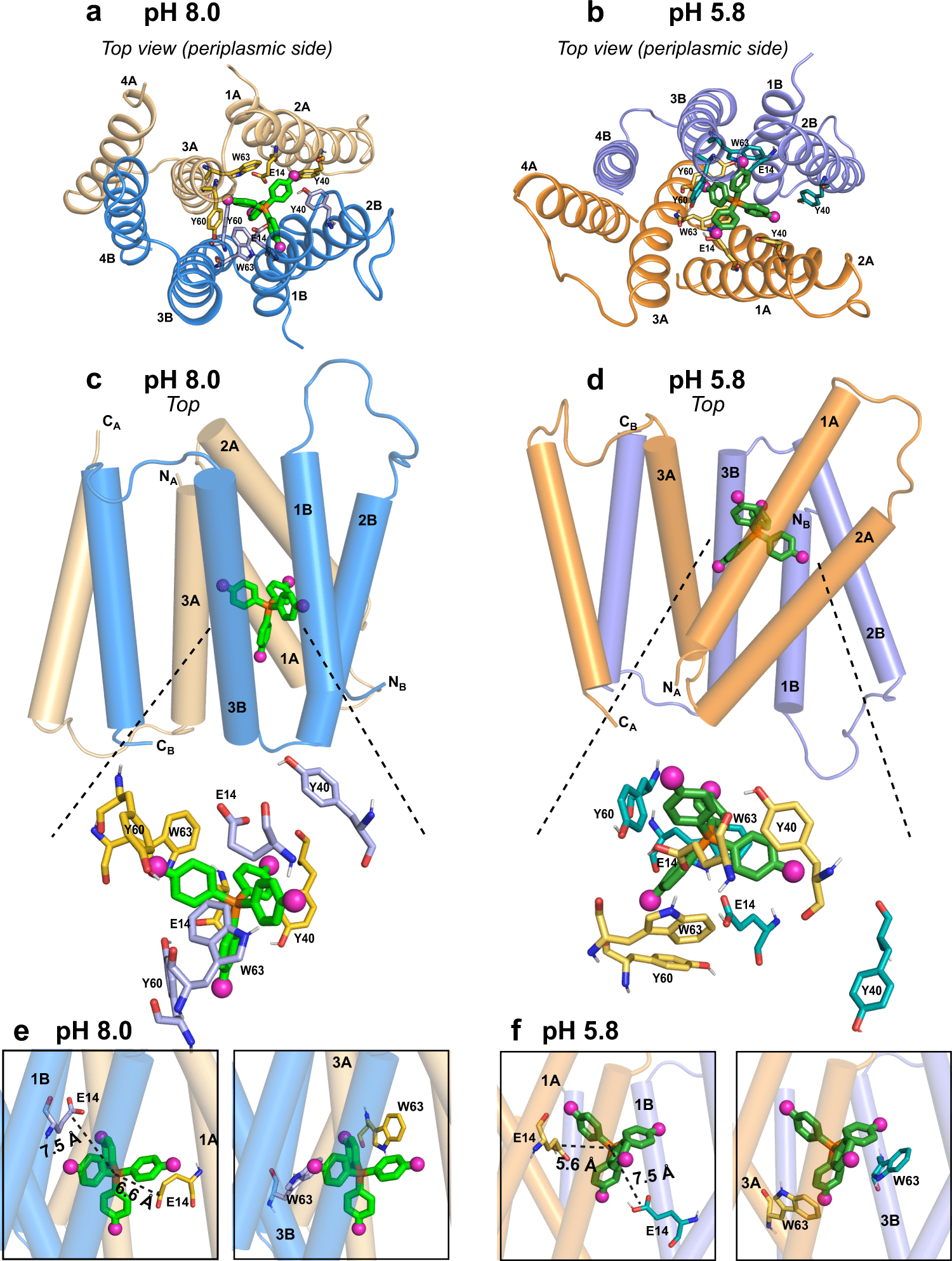
The researchers set out to discover an atomic-resolution structure of EmrE using this ligand and Hong’s innovative NMR approach. Each EmrE molecule has four nearly parallel transmembrane helices, which were previously recognized. Eight transmembrane helices create inner walls that interact with the ligand as it travels through the channel when two EmrE molecules form a dimer. The general architecture of the helices has previously been shown, but not the protein side chains that extend into the channel interior and act as arms to grip the ligand and guide it through the channel.
EmrE is a protein that transfers hazardous chemicals from the neutral pH inside a bacterial cell to the acidic pH outside. The structure of EmrE is affected by the change in pH across the membrane. Hong and Henzler-Wildman found the structure of the protein when it interacts with F4-TPP+ in an acidic environment in a publication published in 2021. They studied the structure at a neutral pH in the new Nature Communications paper, allowing them to see how the structure of the protein changes as the pH varies.
The Full Structure
The channel comprises four helices that are roughly parallel to one another at neutral pH, the researchers discovered in their study, providing an aperture that the ligand may readily enter. The helices begin to tilt when the pH decreases, migrating toward the outside of the membrane, making the channel more open toward the cell’s exterior. This aids in the removal of the ligand from the channel. Several rings in the protein side chains adjust their orientation simultaneously, assisting in the ligand’s exit from the channel.
The channel’s acidic end is likewise more proton-friendly, which enter the channel and aid in its opening, making it easier for the ligand to depart.
“This paper really completes the story,” Hong says. “One structure is not enough. You need two, to figure out how a transporter can actually open to both sides of the membrane, because it’s supposed to pump the ligand or the antibiotic compound from inside the bacteria out of the bacteria.”
Because the EmrE channel is thought to convey a variety of hazardous substances, Hong and her colleagues are currently investigating how other molecules pass through it.
Story Source: Shcherbakov, A.A., Spreacker, P.J., Dregni, A.J. et al. High-pH structure of EmrE reveals the mechanism of proton-coupled substrate transport. Nat Commun 13, 991 (2022). https://doi.org/10.1038/s41467-022-28556-6.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.






Excellent