MarkerGeneBERT, a natural language processing (NLP) system to automatically extract cell type markers from single-cell sequencing literature by parsing full text, was recently introduced by Scientists from CapitalBio Technology, China. MarkerGeneBERT compiled markers for 1733 human and 1832 mouse cell types across tissues, achieving 75% precision and 76% recall. This cell marker gene identification method outperformed CellMarker2.0, suggesting that NLP text mining may be useful for finding cell marker genes.
Introduction
In recent years, a wide range of animals and tissues have undergone the widespread application of single-cell RNA sequencing, or scRNA-seq. The single-cell sequencing technique, known for its high resolution, has paved the way for study in several species and tissues. The main benefit is its ability to accurately map the cellular landscape, including all cell types inside different tissues and organs. To annotate different cell types, researchers must first identify probable cell types in the tissue and then find the matching marker genes through literature studies or existing databases. CellAssign and scCATCH, mechanical tools for coarse-grained annotation, rely on pre-existing or customized cell marker databases.
Although single-cell sequencing has transformed our knowledge of tissues, it is still difficult to distinguish between the many cell types that make up each one. Traditionally, cell markers—genes unique to particular cell types—have been compiled into databases by hand by researchers. This method takes a lot of time, though, and it could overlook fresh findings. Natural language processing, or NLP, can help with this. An NLP system called MarkerGeneBERT was developed by researchers to gather cell marker data from research publications automatically. Consider a relentless AI assistant that examines hundreds of publications, emphasizing crucial details regarding different cell types and their markers. In basic terms, MarkerGeneBERT does that function.
It achieves this through the following steps:
Data Collection:
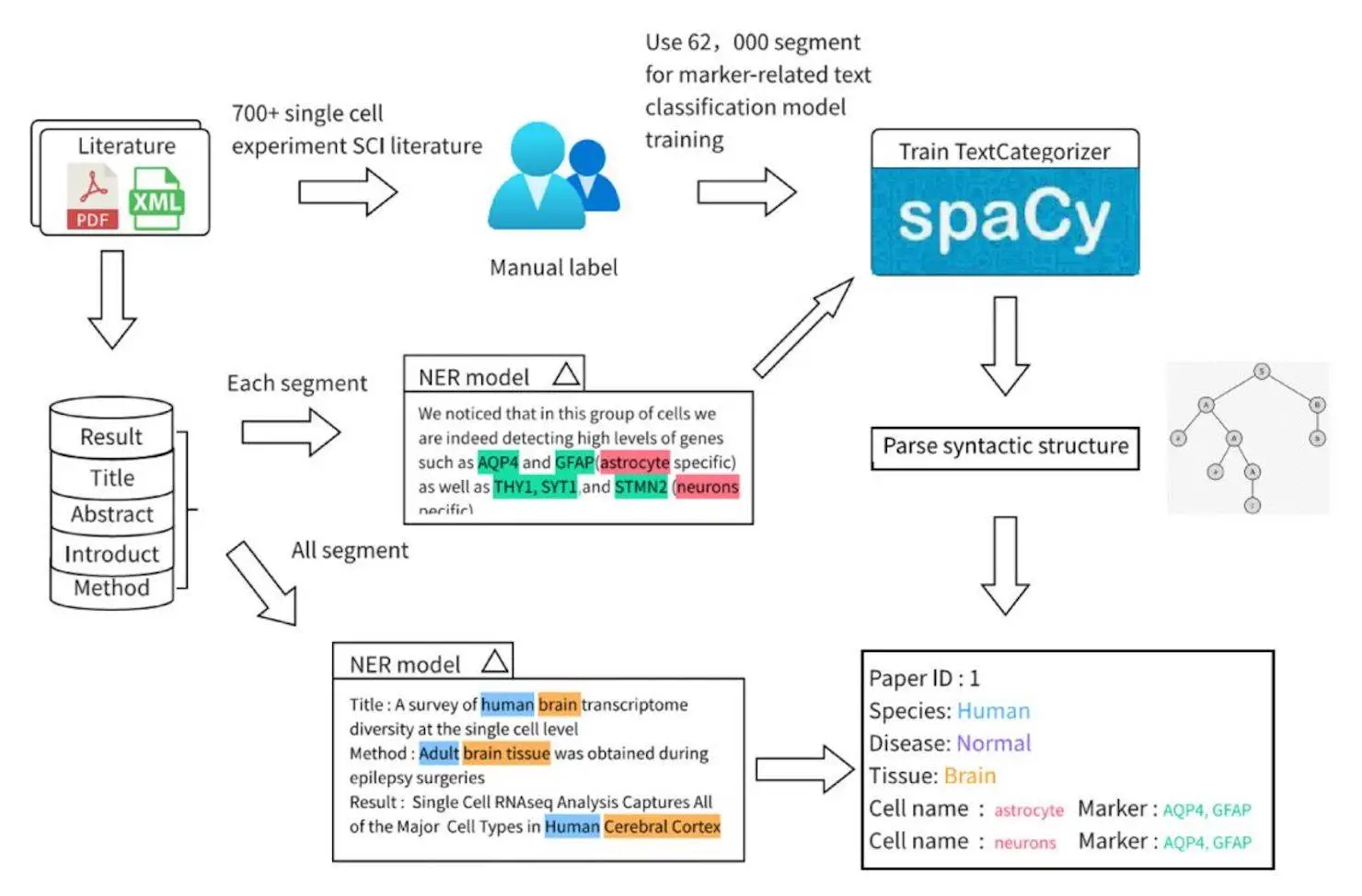
- Researchers have compiled 20,000 research studies on single cells from PubMed and PubMed Central.
- Titles, abstracts, and the body of the material were taken out.
- The introduction, methodology, and results sections—which are specialized—were taken out for closer inspection.
Marker-related Sentence Classification:
- A supervised learning model was trained to detect statements with a certain structure, which included both the name of the cell and the gene in question (for example, “Gene A is a marker of Cell B”).
- Bioinformatics engineers carefully annotated 62,000 sentences.
- Two text preparation approaches were used: lemmatization and part-of-speech tagging.
- A text classification model was trained with spaCy’s TextCategorizer to determine whether or not a sentence is marker-related.
Entity Extraction:
- Named Entity Recognition (NER) models were applied to identify numerous items in the text, comprising cells, species, tissues, and illnesses.
- Multiple models were implemented depending on the strengths of each entity type.
- A full-text-based approach was employed for species and tissue entity recognition, taking into account information from various portions of the manuscript.
Classification of Cell Type-Gene Relation:
- Researchers looked at sentences that had both gene and cell names.
- If a cell-gene relationship existed in the statement, it was predicted using a text classification model.
- From relevant sections, cell-gene pairs were retrieved using rules and syntactic dependency analysis.
The outcomes are remarkable. MarkerGeneBERT found approximately 8,800 cell markers in human tissues and more than 9,000 in mice tissues. This demonstrates an astonishing range of cell kinds. Even more intriguingly, it discovered 183 marker genes and 89 cell types that were absent from earlier databases, offering the prospect of groundbreaking discoveries. However, MarkerGeneBERT is capable of recognizing more than only known markers. It achieved results that were consistent with previous studies by efficiently using its knowledge to annotate brain tissue data. This demonstrates how it may speed up research and save scientists a significant amount of time and effort.
MarkerGeneBERT potential
MarkerGeneBERT illustrates the potential of text data for efficient cell marker extraction. To identify marker genes associated with particular cell types, it analyzes research articles, providing useful data for single-cell RNA sequencing analysis. The approach makes use of supervised learning models trained on carefully annotated data to recognize marker-related sentences within the text. MarkerGeneBERT is capable of communicating the desired connections between genes and cells, even in cases where language is different because of its emphasis on phrase form. This is especially helpful in contrast to co-occurrence-based methods, which might overlook significant signs because of low-frequency co-occurrence.
Key Findings
- MarkerGeneBERT outperformed prior models, obtaining an F1 score of 87% for gene recognition and 92% for cell name identification.
- Over 20,000 genes, as well as 4000 cell types, have been identified from 3987 literature sources.
- When compared with human-curated datasets, MarkerGeneBERT showed outstanding consistency in identifying gene and cell entities, achieving 90.8% and 92.7% accuracy, respectively.
- It also revealed 1764 distinctive cell types that were not previously documented in databases.
- MarkerGeneBERT may be used to annotate cell types from single-cell RNA sequencing data effectively.
There are extensive ramifications. Quicker identification of cell types may help us comprehend illnesses, development, and regeneration more quickly. Furthermore, mass text mining provides access to extensive assessments of gene expression unique to certain cell types in various tissues and animals.
Conclusion
An NLP system named MarkerGeneBERT analyzes research papers to identify marker genes and cell types. Even with distinct terminology, it uses supervised learning models trained on labeled data to identify marker-related phrases. This method outperforms co-occurrence techniques. After obtaining an F1 score of 87% for gene identification and 92% for cell identification, MarkerGeneBERT successfully extracted 20,000 genes and over 4000 cell types from more than 3900 studies. Its limitations, however, are limited to studies on humans and mice, a lack of standard tissue nomenclature, and a concentration on the source text. Future studies will combine various training data sets and use structured graph transformer models to enhance marker-related text classification and entity relationship recognition.
The scientists want to enhance marker-related text classification and entity link identification in the future by employing structured graph transformer models and expanding their training data set. MarkerGeneBERT claims to be a far more effective tool for exposing the secrets buried inside the scientific literature by overcoming these limitations and bringing enhancements, thus speeding up single-cell research.
Article source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.