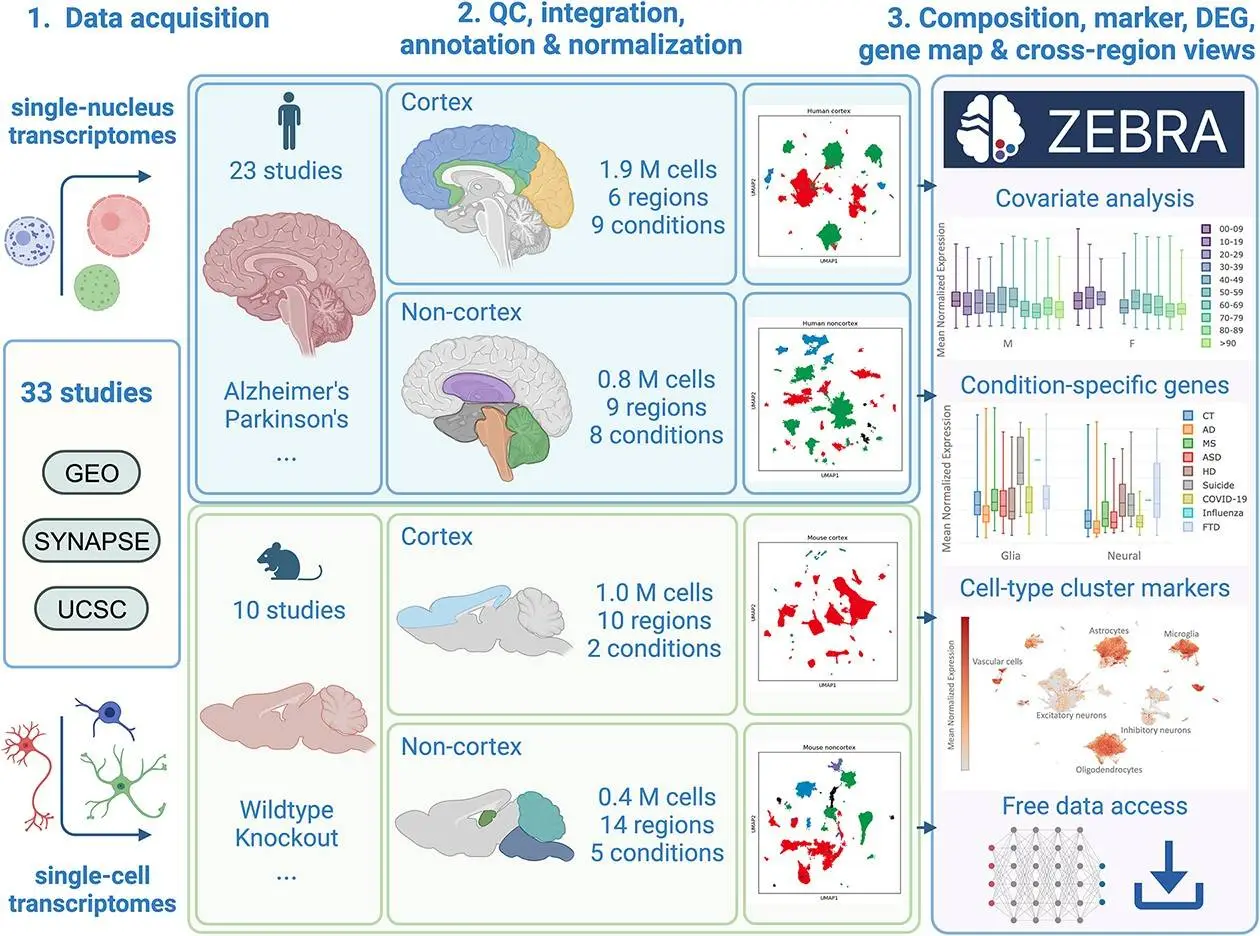
Neurodegenerative diseases like Alzheimer’s and Parkinson’s remain poorly understood despite intensive research. While sequencing studies have profiled diverse brain cell populations, integrating findings into a coherent framework is challenging. To overcome this, researchers from Saarland University, Germany, developed ZEBRA – an expansive database of single-cell and single-nucleus RNA sequencing studies focused on illuminating the molecular underpinnings of neurodegeneration.
The Need to Map Neurodegenerative Transcriptomes
With aging populations, neurodegenerative disorders are rising in prevalence and remain without a cure. These diseases are influenced by genetics, cell type heterogeneity, and other factors. However, the major mechanisms driving pathogenesis are still unknown.
Single-cell transcriptomics enables fine-grained investigation of altered gene expression programs underlying disease. However, translating findings into models of neurodegeneration requires assimilating growing datasets into a unified resource. This has proven difficult due to inconsistent annotations and brain region biases. A comprehensive database is needed to harness published knowledge and drive discovery.
Introducing ZEBRA: A Multi-Study Neurodegeneration Atlas
To address this need, the researchers curated and integrated 33 human and mouse single-cell/nucleus RNA sequencing studies into ZEBRA – the largest neurodegenerative disease transcriptomics resource available. Its key features include:
- 4.2 million cells from human and mouse brain cells sampled from 39 brain regions
- Samples from Alzheimer’s, Parkinson’s, multiple sclerosis patients and models
- Manual curation and normalization using scVI for seamless integration
- Cell type annotation, differential expression, and gene search functionality
By synergizing dispersed data, ZEBRA provides the molecular map to navigate neurodegeneration’s complex terrain and elucidate regulatory mechanisms.
Diving Deep into Brain Cell Populations and Regions
ZEBRA affords rich interactive exploration of molecular profiles across brain cell diversity:
- Visualize cell embeddings colored by type, region, age, or other factors
- Explore expression and differential patterns for specific genes of interest
- Compare cell type composition across conditions using abundance plots
- Find cell type markers and their presence across studies and regions
- Access curated taxonomies of cell definitions across datasets for consistency
- Download data for flexible offline analysis and method development
These capacities empower investigation into cell type-specific vulnerabilities and responses linked to neurodegeneration.
Dissecting Molecular Signatures of Alzheimer’s and Parkinson’s
A key application elucidates transcriptional hallmarks of diseases like Alzheimer’s and Parkinson’s. ZEBRA enables:
- Contrasting patients vs controls to uncover dysregulated genes and pathways
- Stratifying signatures by cell type to pinpoint the most affected populations
- Identifying common mechanisms across mouse models and human patients
- Comparing related diseases like Alzheimer’s and multiple sclerosis for shared patterns
- Exploring gene expression changes across the age spectrum for insights into aging
Such analytical workflows can drive hypothesis generation and biomarker discovery for neurodegenerative diseases.
The Future of Integrative Transcriptomics
By providing an invaluable translational scaffold, ZEBRA advances our understanding of neurodegeneration’s complex molecular fabric. In future, the researchers intend to focus on:
- Expanding integrated data spanning additional studies, tissues, and species
- Aligning raw reads for improved resolution of splicing and isoform diversity
- Multi-omics integration to connect transcripts with proteins, metabolites, and genetics
- Improved taxonomies and ontologies for greater standardization
- Tools for network analysis and trajectory inference from sc/snRNA-seq data
- Clinical applications from patient stratification to therapeutic development
As single-cell techniques continue advancing, ZEBRA represents the next phase in maximizing their potential – aggregating isolated insights into unified knowledge to cure devastating neurological diseases.
Conclusion
ZEBRA exemplifies the power of synergizing distributed data into cohesive models of biology. By providing an invaluable neurodegeneration transcriptomics resource, it aims to unravel the intricate mechanisms driving dementia, Parkinson’s, Multiple Sclerosis, and related disorders. The researchers envision ZEBRA accelerating the development of urgently needed diagnostics and therapies to tackle these diseases and illuminate the complex functions of the enigmatic brain.
Article Source: Reference Paper | ZEBRA is available at the Website
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.