A platform for separation-free detection of bacteria in arbitrary medium has been developed by the researchers from Korea Advanced Institute of Science and Technology (KAIST), using a synergistic integration of surface-enhanced Raman spectroscopy and deep learning.
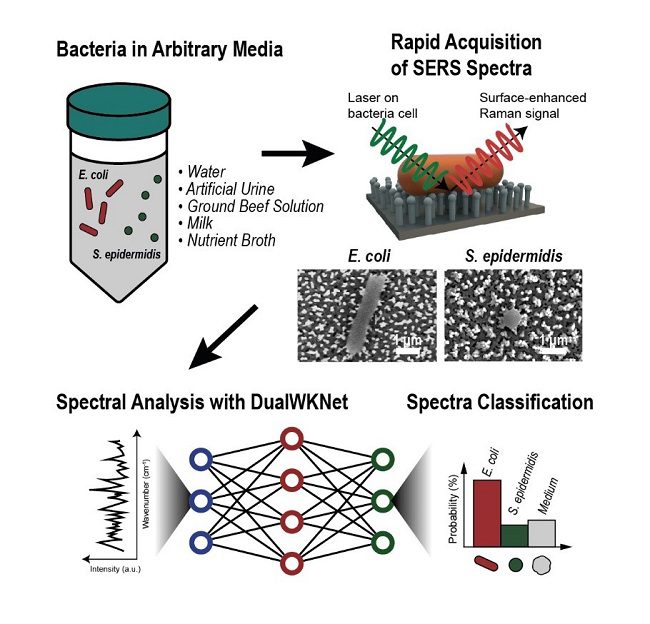
Image Source: https://news.kaist.ac.kr/newsen/html/news/?mode=V&mng_no=19210&skey=&sval=&list_s_date=&list_e_date=&GotoPage=1
Bacterial identification can take hours, if not days, to complete, wasting valuable time for diagnosing diseases and choosing treatments. According to KAIST experts, there may be a faster and more precise method. The researchers were able to categorize bacteria in diverse environments with 98 percent accuracy by teaching a deep learning system to recognize the “fingerprint” spectra of the chemical components of individual bacteria.
The findings were published online in Biosensors and Bioelectronics on Jan. 18 ahead of print in the journal’s April edition.
Bacteria-induced illnesses, such as those caused by direct bacterial infection or exposure to bacterial toxins, can cause painful symptoms and even death, so rapid detection of bacteria is essential for avoiding contaminated food consumption and diagnosing infections from clinical samples like urine. “We demonstrated a markedly simple, fast, and effective route to classify the signals of two common bacteria and their resident media without any separation procedures by using surface-enhanced Raman spectroscopy (SERS) analysis boosted with a newly proposed deep learning model,” told Professor Sungho Jo of the School of Computing.
Raman spectroscopy examines how light scatters across a substance. The spectral fingerprint reveals structural information about the material, allowing researchers to identify its constituents. Sample cells are placed on noble metal nanostructures in the surface-enhanced version, which helps amplify the sample’s signals.
Due to various overlapping peak sources, such as proteins in cell walls, it is difficult to get consistent and unambiguous spectra of bacteria. “Moreover, strong signals from the surrounding medium are amplified to overpower target signals, necessitating time-consuming and labor-intensive bacterial separation operations,” said Professor Yeon Sik Jung of the Department of Materials Science and Engineering.
The researchers used deep learning, an artificial intelligence technology that can hierarchically extract particular elements of spectral information to categorise data, to filter through the noisy signals. Their approach, called the dual-branch wide-kernel network (DualWKNet), was created expressly to understand the link between spectral properties quickly. According to Professor Jo, this capacity is essential for interpreting one-dimensional spectral data.
“Despite having interfering signals or noise from the media, which make the general shapes of different bacterial spectra and their residing media signals look similar, high classification accuracies of bacterial types and their media were achieved,” Professor Jo said, explaining that DualWKNet allowed the team to identify key peaks in each class that was almost indiscernible in individual spectra, enhancing the classification accuracies. “Ultimately, with the use of DualWKNet replacing the bacteria and media separation steps, our method dramatically reduces analysis time.”
The researchers intend to utilize their platform to investigate more bacteria and media kinds, then use the data to create a training data library of numerous bacterial varieties in more medium to reduce the time spent in collecting and detecting fresh samples.
With the use of SERS and deep learning, the researchers were able to create a useful universal platform for quick bacterial identification. The researchers are further trying to extend the usage of a deep learning-based SERS analysis platform to detect a variety of bacteria in new media, such as blood, that are relevant for food or clinical study.
Story Source: Eojin Rho, Minjoon Kim, Seunghee H. Cho, Bongjae Choi, Hyungjoon Park, Hanhwi Jang, Yeon Sik Jung, Sungho Jo, “Separation-free bacterial identification in arbitrary media via deep
neural network-based SERS analysis,” Biosensors and Bioelectronics online January 18, 2022 (doi.org/10.1016/j.bios.2022.113991)
https://news.kaist.ac.kr/newsen/html/news/?mode=V&mng_no=19210&skey=&sval=&list_s_date=&list_e_date=&GotoPage=1
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.





