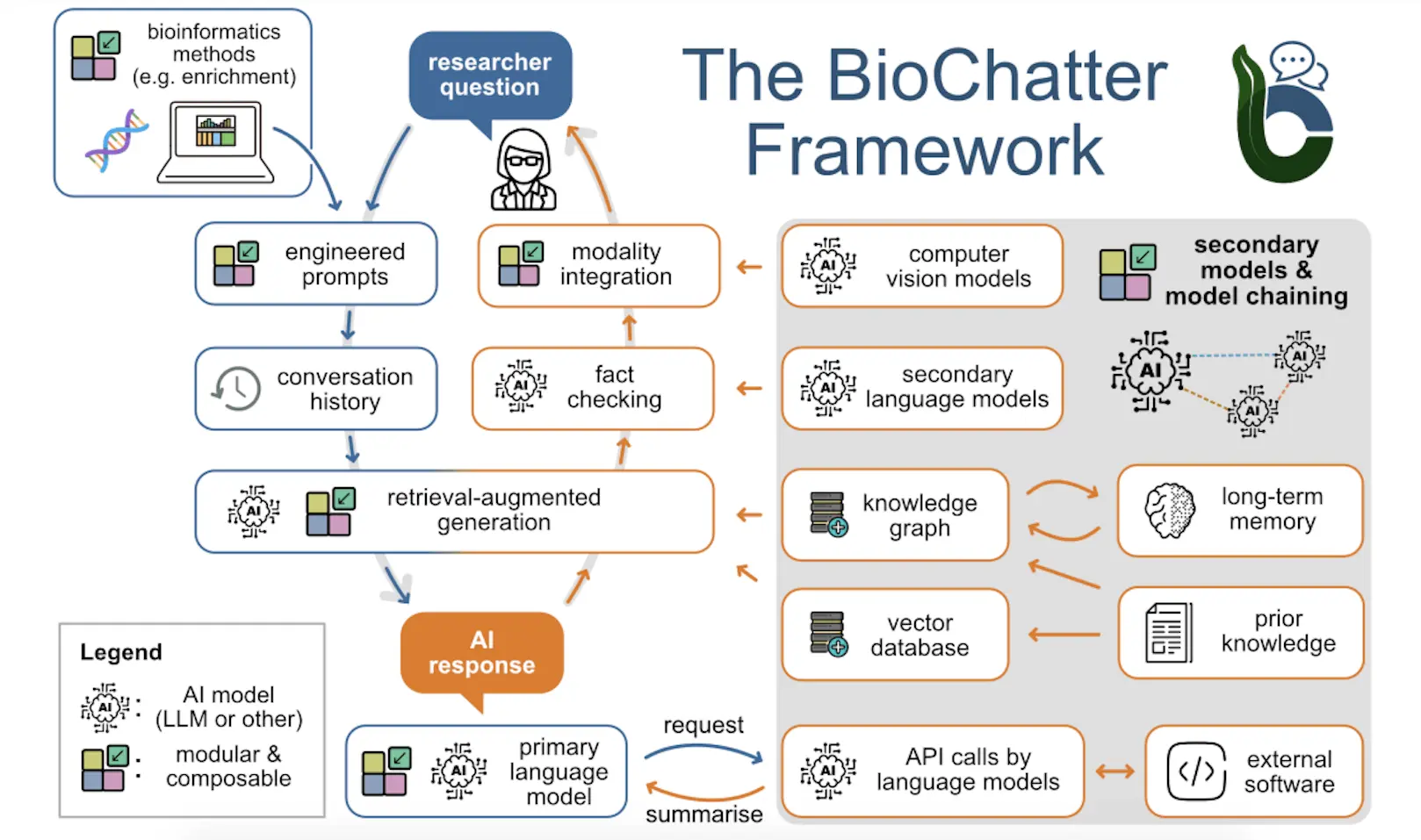
BioChatter, a new open-source framework developed by Heidelberg University researchers and collaborators, aims to harness the power of large language models (LLMs) for biomedical research while ensuring responsible use. By integrating various functionalities like knowledge retrieval and model chaining, BioChatter simplifies LLM interaction for researchers, making it accessible and inclusive. It also emphasizes user-friendliness and privacy, allowing for the deployment of local LLMs that protect sensitive data. This framework has the potential to revolutionize how we utilize LLMs for biomedical discovery and development.
What Are Large Language Models and Why Do They Matter for Biomedicine?
Understanding complex biological and biomedical systems poses major challenges despite technological advances that allow us to collect vast amounts of data. Even domain experts cannot fully know the implications of every gene, molecule, symptom, or biomarker due to the inherent limitations of human knowledge. Biological events are also highly context-dependent based on factors like cell type or disease.
In contrast, large language models of the current generation, like GPT-3, can access enormous amounts of knowledge encoded in their parameters. If trained correctly, they can recall and combine virtually limitless knowledge from their training set. LLMs have taken the world by storm, and many biomedical researchers already use them in their daily work to assist with both general and research tasks.
However, interacting with LLMs is mostly a manual, non-reproducible process, and their behavior can be unpredictable or erratic at times. For example, they are prone to confabulation, where they make up convincing but false facts. More oversight is needed when applying LLMs in sensitive biomedical contexts related to data privacy, licensing, transparency, and ethics.
Introducing BioChatter – An Optimized Framework for Biomedicine
To facilitate interfacing with LLMs for biomedical research while constraining their functionality for sensible and ethical application, the researchers developed BioChatter. It is an open-source framework optimized for communicating with LLMs about biomedical tasks like experimental design, outcome interpretation, literature review, and resource exploration.
BioChatter guides users intuitively through interacting with models while counteracting problematic LLM behaviors. It focuses on robust and user-friendly implementation, including the option to run privacy-preserving local open-source LLMs. The modular design means components can be swapped out and relies on integrating existing open-source software related to LLMs.
Key Framework Functionalities
The core functionality of BioChatter is a question answering and connectivity with both leading proprietary models like GPT-3 and open-source alternatives such as LLaMA or Mixtral. OpenAI models can be accessed via API to improve data privacy protections compared to the web interface.
The researchers emphasize supporting open-source LLMs for greater transparency and the ability to run models locally to avoid privacy concerns of cloud-based services. The design allows easy exchange of models to keep up with the rapid evolution of open-source LLMs.
- Prompt Engineering
LLMs are highly sensitive to the initial prompt guiding them to a specific behavior or task. However, current prompt engineering practices lack reproducibility and must be re-engineered for each new model.
BioChatter includes a prompt framework to preserve sets for particular tasks that can be shared and reused. Automated benchmarking integrates prompts to evaluate their performance across models.
- Knowledge Graphs
Knowledge graphs (KGs) represent information in a structured, queryable way. The BioCypher framework creates KGs from biomedical data with ontological grounding for context. BioChatter builds upon this by enabling natural language interactions with BioCypher KGs.
The KG build information is used to tune LLM understanding for more efficient querying. Connecting to BioCypher KGs also allows integrating prior knowledge to ground LLM responses.
- Retrieval-Augmented Generation
To address LLM confabulation issues, retrieval-augmented generation (RAG) injects relevant information into prompts without expensive retraining. While KGs can provide structured knowledge, retrieving unstructured data like literature is often more efficient.
By managing vector databases, BioChatter users can embed documents for semantic search to augment prompts with relevant text fragments. This improves factual accuracy by grounding responses in documents.
- Model Chaining and Fact Checking
LLMs can orchestrate multi-step tasks by interacting with other models and APIs. However, implementation complexity introduces unpredictability. BioChatter develops tailored solutions focused on stability for common biomedical applications like experimental design and literature analysis.
For example, the researchers implemented a fact-checking module using a secondary LLM to continually assess the primary LLM’s response accuracy during conversations.
- Benchmarking
The increasing generality of LLMs makes comprehensive evaluation challenging. To address this, the authors focused on biomedical tasks and datasets with automated validation of responses. Transparent and reproducible comparisons rely on a benchmarking framework to evaluate combinations of models, prompts, and other pipeline components.
Encryption is implemented to prevent benchmark data from leaking into LLM training sets. The benchmark enables convenient model selection and flags issues like the performance decline between GPT-3 versions. The analysis confirmed OpenAI’s leading performance, but some open-source models reached high task-specific scores.
Responsible Development and Adoption
The rapid pace of LLM progress challenges society and biomedicine to integrate these powerful technologies responsibly. Their application requires expertise many researchers lack. Collaboration is also hampered by the complexity of the field and systemic incentives against open science.
Inspired by open-source libraries like LangChain, the researchers designed BioChatter as an inclusive framework focused on application rather than engineering challenges. They reused existing open-source tools and adapted recent LLM advancements for the biomedical context through ongoing transparency.
Efficient human-AI interaction may require a “lingua franca” with symbolic representation of concepts. BioChatter enables this by grounding interactions in BioCypher KGs mapped to standard ontologies and terminology for contextual shared understanding between the user and LLM.
The researchers emphasize evaluating LLM behavior through living benchmarks, assessing components automatically across tasks relevant to Biomedical applications. Ongoing encryption prevents benchmark data from influencing new models.
So far, GPT models lead overall, but open-source alternatives have reached high performance for particular tasks. Benchmarking also quickly flagged GPT version performance declines for further investigation. Manually intensive small-scale benchmarks reliant on opaque web interfaces obscuring key parameters were avoided.
Access flexibility allows proprietary models to provide economical access alongside open-source options to catch up rapidly to sustain long-term progress. The deployment framework hosts models at any scale, from dedicated GPUs to user laptops or browsers.
Despite efforts to constrain functionality safely, LLMs still carry risks if applied unchecked in biomedicine’s diverse subfields. LLMs can be seen as assistants enhancing human strengths rather than autonomous replacements needing oversight. Adoption may still encounter learning curves for those unfamiliar with LLMs or BioChatter specifically.
As research on abilities like multimodal learning and agent behaviors matures, BioChatter can integrate advances beneficially while continually encouraging open-source transparency. The framework’s concepts could also extend to other scientific fields by adjusting domain-specific inputs in a user-friendly way.
Conclusion
BioChatter enables biomedical researchers to harness the promise of large language models efficiently through purpose-built constraints and functionalities targeted to realistic use cases. Ongoing collaborative and transparent development focused on robust evaluation aims to further improve human-AI symbiosis in biomedicine while avoiding pitfalls from uncontrolled application.
Article source: Reference Paper | BioChatter framework is available on GitHub | BioChatter: Web Apps
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.