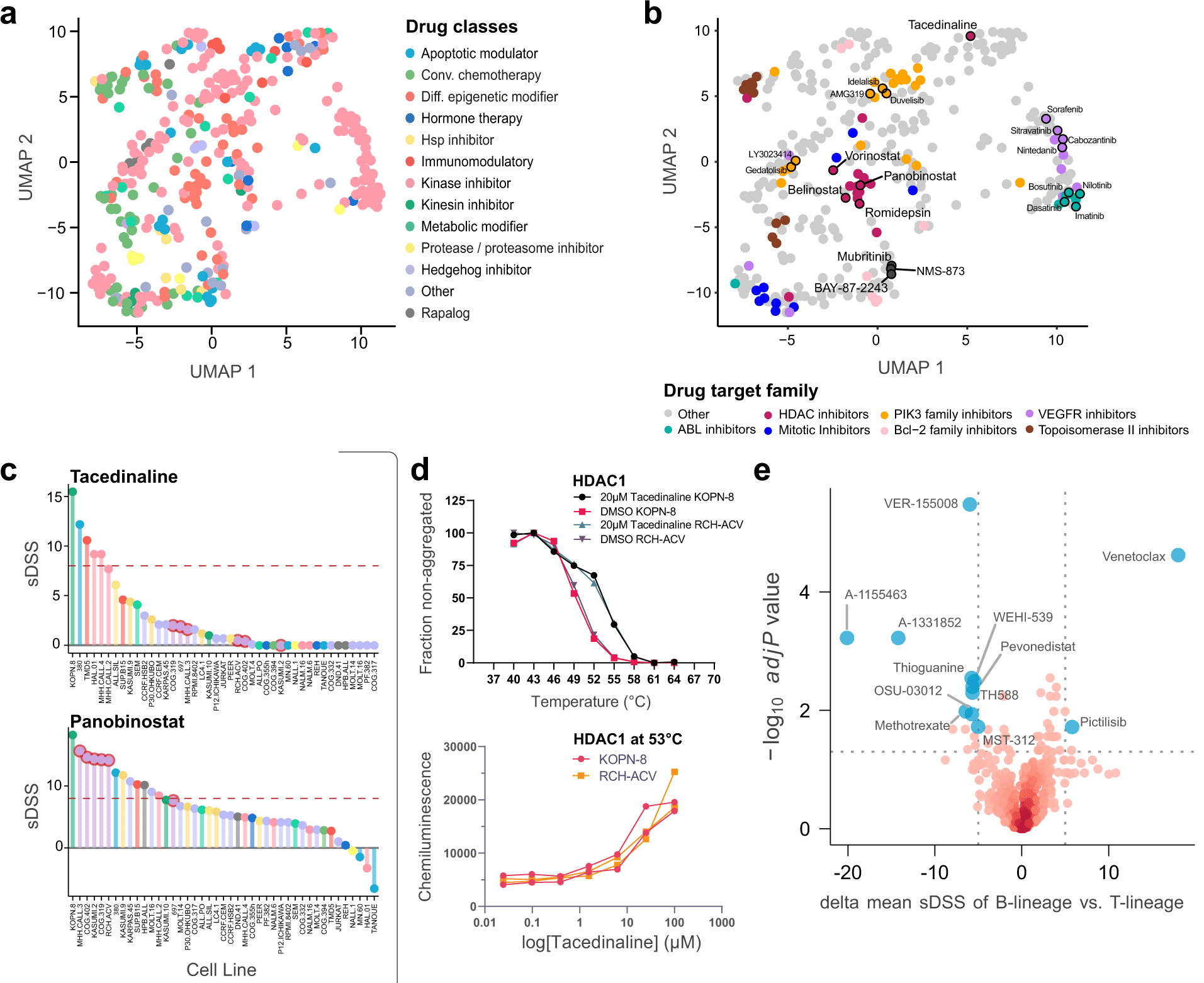
In a report published in Nature Communications, researchers from the Department of Oncology-Pathology at Karolinska Institutet, Sweden established a cell line resource dataset and analysis tool ‘Functional Omics Resource of ALL (FORALL)’ that could be used to uncover the features that make specific leukemias vulnerable to new forms of targeted medicines.
Image Source: Integrative multi-omics and drug response profiling of childhood acute lymphoblastic leukemia cell lines.
The researchers used proteomics, transcriptomics, and pharmacoproteomic characterization to perform comprehensive multi-omic studies of 49 widely available childhood acute lymphoblastic leukemia (ALL) cell lines. The drug associations, as well as lineage-dependent correlations, were found by connecting molecular characteristics with pharmacological responses to 528 cancer medicines.
Comparing cell lines that respond to medications in different ways could help researchers determine how drugs work and what characteristics make leukemias susceptible or resistant to them. The researchers discovered that some medicines might function through secondary off-target processes. They found surprising sensitivity to a form of immunotherapy medicine that strengthens the molecular signals that activate immune cells in a higher-risk ALL subtype that responds poorly to regular chemotherapy.
ALL is the most frequent cancer in children. Most patients can be treated with chemotherapy and stem cell transplantation, but 15-20% will not respond well, leaving them few treatment options and a significant risk of death. Furthermore, current standard therapies can have long-term health consequences for children who survive cancer. There is a requirement to find less harsh therapies than standard treatments that can aid patients who aren’t responding to present medications. The findings of the researchers serve as a foundation for further research into biological distinctions and potential targeted therapies in children with ALL, as well as a large public resource to support other research efforts.
Image Source: Integrative multi-omics and drug response profiling of childhood acute lymphoblastic leukemia cell lines.
Using mass spectrometry, the researchers employed a panel of 49 childhood ALL cell lines to perform in-depth proteome measurements, RNA sequencing, high-throughput drug screening, and fusion gene analyses. They built a web-based platform that allows anyone to browse and analyze the data. They ran analyses that combined different data sets and helped them better comprehend biological distinctions. They ran tests to profile the mechanisms of the most effective medications on the screen. They carried out validation tests to have a better understanding of pharmacological processes.
According to Rozbeh Jafari, a researcher at the Department of Oncology-Pathology and the paper’s senior author,” the researchers will further validate the results in the dataset with the highest potential for future clinical benefit.” The researchers want to expand the number of juvenile leukemia subtypes represented in the dataset, as well as to characterize the drug sensitivity and biological mechanism findings in clinical samples and additional models. The researchers are also interested in establishing new ways to analyze and interpret large datasets that measure various molecular characteristics and developing new approaches to understanding the pharmacological activity.
Final thoughts
In this study, the authors used proteomics, transcriptomics, and pharmacoproteomics (multi-omics) to analyze 49 childhood acute lymphoblastic leukemia cell lines. The researchers found that the diacylglycerol-analog bryostatin-1 was a therapeutic candidate in the MEF2D-HNRNPUL1 fusion high-risk subtype, for which this drug activates pro-apoptotic ERK signaling associated with molecular mediators of pre-B cell negative selection.
Article Sources: Reference Paper | Reference Article |Interactive Shiny App | GitHub Repository
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.