Scientists used neutrons to learn more about how aurein 1.2, an antimicrobial peptide (AMP) that attacks bacteria's membranes, kills antibiotic-resistant bacteria. The research will be crucial in identifying and developing more efficient and cost-effective AMPs.
Antibiotic drugs have saved countless lives by preventing and treating bacterial infections since the discovery of penicillin more than 90 years ago. Nevertheless, bacteria are rapidly developing new ways to resist antibiotics, rendering some of modern medicine’s most potent drugs ineffective against a wide range of life-threatening infections, including gonorrhea, and Methicillin-resistant Staphylococcus aureus (MRSA), and Clostridioides difficile (C. difficile), to name a few. In the United States alone, the Centers for Disease Control and Prevention (CDC) estimates that about 3 million drug-resistant illnesses occur each year.
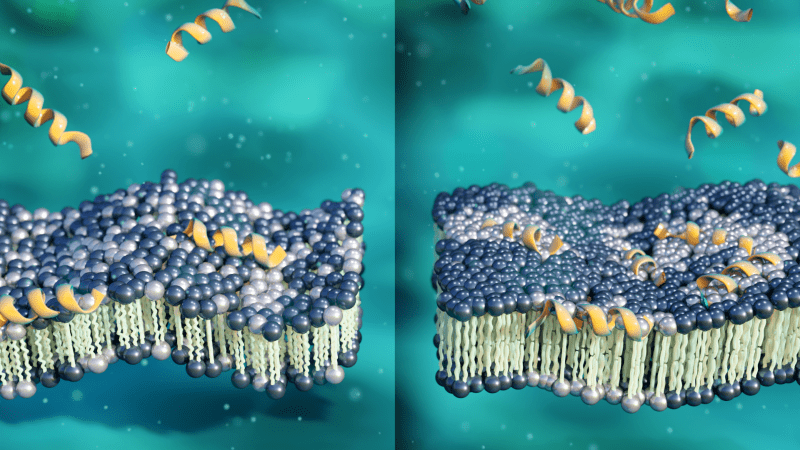
Scientists used neutrons to better understand how an antimicrobial peptide known as aurein 1.2 kills antibiotic resistant bacteria by attacking their membranes. As the peptide concentration increases (yellow), aurein 1.2 induces lipid molecule clusters (white) within the membrane, making it thicker, more rigid, and more susceptible to break down under stress. Their findings could inform new strategies for treating infections where antibiotics have fallen short. (Credit: ORNL/Jill Hemman).
This surge in resistance has prompted a frantic quest for new ways to combat bacterial infections. Developing peptides, which are strings of amino acids with a propensity for neutralizing bacterial invaders, is one possible technique for tackling antibiotic resistance.
Antimicrobial peptides (AMPs) are peptides that act as an intrinsic defense against microbial infections in animals, plants, and other multicellular organisms. These peptides are regarded as promising therapeutic possibilities for treating bacterial infections when antibiotics have failed because of their ability to identify and kill a wide spectrum of microorganisms promptly.
Neutron experiments undertaken by scientists at the Department of Energy’s Oak Ridge National Laboratory (ORNL) have revealed new information about how AMPs prevent bacterial infections. The team put together the molecular physics behind an effective AMP termed aurein 1.2’s capacity to cause severe damage to bacterial cells in small amounts by examining it. Their findings, which were reported in BBA Advances, could aid pharmaceutical companies in developing more efficient and effective antibiotics to combat antibiotic-resistant bacteria.
According to the leading author of the research Shuo Qian, a neutron scattering scientist at ORNL, AMPs attack bacteria by using electrostatic and hydrophobic forces to target their membranes. A cell membrane’s basic structure is a double layer of lipid molecules. The negatively charged lipid molecules that make up the surface of bacterial cell membranes are very attracted to positively charged AMPs. The hydrophobic peptides will commonly lodge themselves into the membrane once bound to an entering bacterial cell to avoid exposure to water.
AMPs bore deep holes in bacterial membranes when they were present in large enough numbers. This activity weakens the invading bacterial cells by disrupting the membrane’s structure. The disadvantage is that creating medicines with high peptide concentrations could be more expensive and potentially cause patient adverse effects. Drugs that destroy antibiotic-resistant bacteria without requiring a high peptide count might be better for patient health and less expensive. This is when aurein 1.2 came in handy for resolving the problem.
Aurein 1.2, which is only 13 amino acids long, is one of the shortest AMPs found in nature. Synthetically created copies of this peptide, which was first found in Australian tree frogs, have shown to be effective against a number of bacteria. The chemical mechanism behind its pathogen-fighting properties, on the other hand, is mainly unclear.
Because neutrons can non-destructively examine the chemical constituents that make up biological systems, such as peptides and membranes, neutron scattering is an excellent approach for gathering extensive information on them.
Previous neutron tests led by Qian at ORNL, which were reported in Scientific Reports and Langmuir, revealed some preliminary insights into the peptide’s mysterious mechanism. Aurein 1.2, instead of puncturing holes in bacterial membranes as other AMPs do, interferes with the membranes’ electrostatic balances, according to these findings. The peptide’s positive charge causes lipid molecules to slow down and form clusters with several other similarly charged lipid molecules within the membrane at low concentrations. This was the first time this behavior had been observed during previous AMP interactions.
Qian in his most recent study, collaborated with Piotr Zolnierczuk, an ORNL neutron scattering expert, to better understand how peptide-induced lipid clusters affect bacterial membrane function. At ORNL’s Spallation Neutron Source, they used two neutron scattering devices (SNS).
Qian discovered that aurein 1.2 remained entrenched near the membrane’s surface even at high concentrations using the EQ-SANS equipment to detect peptide penetration within the membrane. The impact of aurein 1.2 on membrane flexibility and compressibility was then investigated by Zolnierczuk using the Neutron Spin Echo Spectrometer.
As explained by Zolnierczuk, membrane fluctuations can last tens or hundreds of nanoseconds, and neutron spin echo spectroscopy is the sole technology that can measure dynamical behaviors on that timescale. The information about a membrane’s mechanical properties can be deduced, such as how soft or stiff it becomes, by examining neutron spin echo data using mathematical models.
The research demonstrated that when the membrane is exposed to low peptide concentrations, it thins and becomes more flexible. The peptides cause the membrane to become thicker and stiffer at somewhat greater doses. The findings imply that aurein 1.2 peptides can thicken and rigidify bacterial membranes via lipid-clustering.
In normal conditions, the lipid molecules that make up membranes are constantly moving around to accommodate biological activities. It’s a very fluid, inhomogeneous environment in a delicate balance. However, we found that when aurein 1.2 causes lipid molecules to slow down and form unexpected clumps, the membranes end up becoming more rigid than usual.
Shuo Qian
The bacterial membrane can’t recover as rapidly from stressors, its structural integrity is reduced, and it’s less able to carry out regular duties in this state. The bacterial cell is more prone to break down under stress as a result of this combination of negative impacts. It’s comparable to how freezing temperatures cause water molecules to slow down and flow water to become hard brittle ice.
The findings of this study will aid scientists in better understanding the complexities of AMP-based antibacterial defenses and may impact future therapeutic options for infections in an increasingly antibiotic-resistant environment.
The Second Target Station (STS), which is now under development at ORNL, will considerably increase this type of study in the future. The STS will enable advances in many materials research and development fields by providing radical new capabilities for discovery science.
CENTAUR, one of the first STS instruments, will provide unprecedented capabilities for studying materials with high resolution over a wide range of length scales, measuring smaller samples, and conducting time-resolved investigations of evolving structures in a variety of scientific fields, including biological membranes and other forms of soft matter.
Story Source: Qian, S., & Zolnierczuk, P. A. (2022). Interaction of a short antimicrobial peptide on charged lipid bilayer: A case study on aurein 1.2 peptide. BBA Advances, 2, 100045.
https://neutrons.ornl.gov/content/mystery-mechanism-small-peptide-shows-big-promise-fighting-antibiotic-resistant-bacteria
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.