Scientists from MIT probed into how proteins recognize and bind to one another, informing drug treatments for cancer.
A single cell, despite its small size, has billions of molecules that buzz around and bond to one another, performing critical tasks. About 20,000 proteins are encoded in the human genome, the majority of which interact with partner proteins to mediate over 400,000 different interactions. These companions don’t just attach to each other at random; they only bind to extremely particular friends that they must identify within the overcrowded cell. Cancer or other illnesses might result if they make the incorrect connections — or even the good pairings at the wrong location or time. Scientists are hard at work trying to figure out how these protein-protein interactions function in order to develop medications that disrupt or mimic them to cure illness.
The average human protein is made up of about 400 amino acids, which are linked together and folded into a complicated three-dimensional structure. Some proteins include short linear motifs (SLiMs), which are regions of four to six amino acids that influence protein-protein interactions, inside this lengthy string of building blocks. SLiMs and their binding partners help crucial cellular activities despite their simplicity and modest size. Experiments to investigate how SLiMs detect their unique binding partners have proven problematic in the past.
To solve this difficulty, a team led by Theresa Hwang devised a screening approach to learn how SLiMs selectively attach to certain proteins and can even differentiate between proteins with similar structures. The researchers built their own synthetic chemical capable of attaching particularly firmly to a protein called ENAH, which is linked to cancer metastasis, using the extensive knowledge acquired from examining these interactions. The findings were published in two eLife articles, the first one on Dec. 02, 2021, and the other on Jan. 25, 2022.
“The ability to test hundreds of thousands of potential SLiMs for binding provides a powerful tool to explore why proteins prefer specific SLiM partners over others,” says Amy Keating, the senior author of both papers. “As we gain an understanding of the tricks that a protein uses to select its partners, we can apply these in protein design to make our own binders to modulate protein function for research or therapeutic purposes.”
Generally, SLiM screens currently in use only look for short, tight binders, ignoring SLiMs that don’t hold their partner proteins as tightly. Keating, Hwang, and their colleagues created their own screen, MassTitr, to examine SLiMs with a wide variety of binding affinities.
The researchers also speculated that the amino acids on each side of the SLiM’s core four-to-six amino acid sequence may have an unappreciated role in binding. To put their idea to the test, they utilized MassTitr to screen the human proteome in 36-amino-acid chunks to determine which “extended” SLiMs might bind to the protein ENAH.
ENAH, also known as Mena, is a protein that aids cell movement. Healthy cells require the capacity to migrate, while malignant cells can use it to spread. Scientists discovered that lowering ENAH levels reduces a cancer cell’s capacity to infiltrate other tissues, implying that medications that disrupt this protein and its relationships might be used to treat cancer.
The scientists discovered 33 SLiM-containing proteins that bound to ENAH using MassTitr, 19 of which are possibly new binding partners. They also uncovered three unique amino acid patterns surrounding core SLiM sequences that helped the SLiMs attach to ENAH even more tightly. One of these extended SLiMs, located in a protein called PCARE, has the greatest known affinity for ENAH of any SLiM discovered to date.
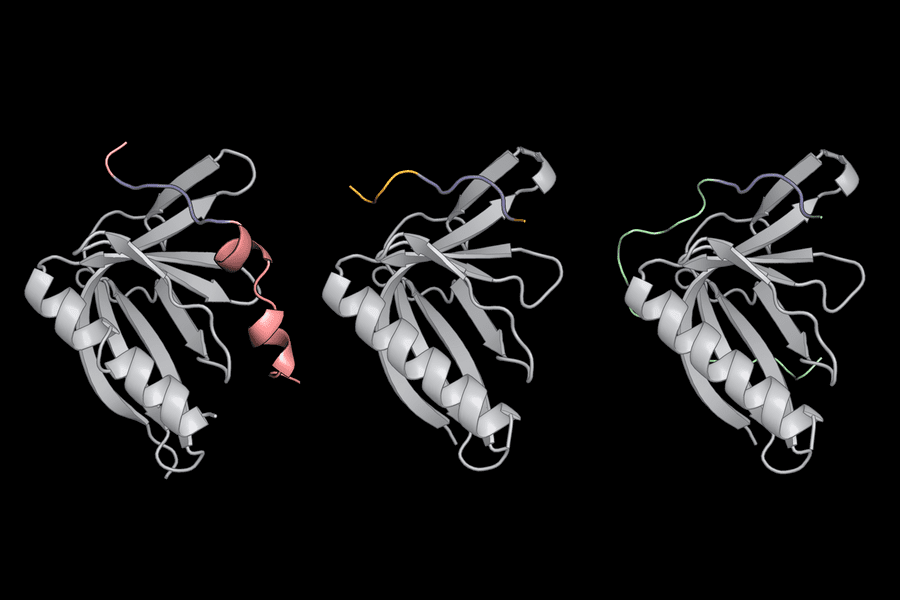
The researchers then used computer software called dTERMen in conjunction with X-ray crystallography to figure out why PCARE attaches to ENAH rather than ENAH’s two virtually similar sister proteins (VASP and EVL). The amino acids surrounding PCARE’s core SliM led ENAH to shift shape somewhat when the two came into contact, allowing the binding sites to grasp onto one another, according to Hwang and her colleagues. Because VASP and EVL were unable to undergo this structural alteration, the PCARE SliM could not bind to them as firmly.
This unique interaction encouraged Hwang to design a protein of her own that binds to ENAH with remarkable affinity and specificity. “It was exciting that we were able to come up with such a specific binder,” says Hwang. “This work lays the foundation for designing synthetic molecules with the potential to disrupt protein-protein interactions that cause disease — or to help scientists learn more about ENAH and other SLiM-binding proteins.”
Ylva Ivarsson, a biochemistry professor at Uppsala University who was not involved in the research, argues that knowing how proteins identify their binding partners is crucial to cell function and control. She notes that the two eLife experiments demonstrate that prolonged SLiMs play an unrewarded role in shaping the affinity and specificity of binding interactions.
“The studies shed light on the idea that context matters, and provide a screening strategy for a variety of context-dependent binding interactions,” she says. “Hwang and co-authors have created valuable tools for dissecting the cellular function of proteins and their binding partners. Their approach could even inspire ENAH-specific inhibitors for therapeutic purposes.”
The experiment taught Hwang that things aren’t always what they seem: even small, basic protein segments may play complicated roles in the cell. “We should really appreciate SLiMs more,” she says.
Story Source: Hwang, T., Parker, S. S., Hill, S. M., Grant, R. A., Ilunga, M. W., Sivaraman, V., … & Keating, A. E. (2022). Native proline-rich motifs exploit sequence context to target actin-remodeling Ena/VASP protein ENAH. eLife, 11, e70680.
Hwang, T., Parker, S. S., Hill, S. M., Ilunga, M. W., Grant, R. A., Mouneimne, G., & Keating, A. E. (2021). A distributed residue network permits conformational binding specificity in a conserved family of actin remodelers. Elife, 10, e70601.
Background Image: Short linear motifs (blue) use flanking regions outside their core amino acid sequence to recognize proteins such as ENAH (gray).Credits:Image: Theresa Hwang. https://news.mit.edu/2022/probing-how-proteins-pair-inside-cells-0203
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.