Scientists from the University of Washington, the Max Planck Institute for Dynamics and Self-Organization, and the University of Cologne carefully formulated combination therapies from broadly neutralizing antibodies (bNAbs) which may aid in the treatment of HIV while reducing the possibility that the virus will evolve to “escape” treatment. The study published in the journal eLife offers new hope for HIV therapy.
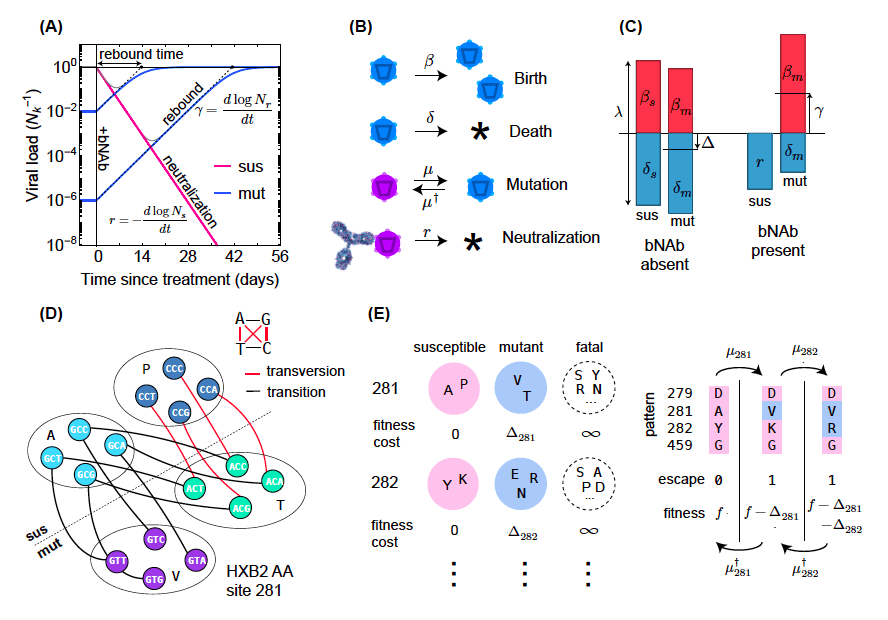
Image Source: https://elifesciences.org/articles/76004
Broadly neutralizing antibodies have the potential to replace anti-retroviral therapy in the treatment of HIV. Suppressing viral escape is a major problem that is easier to accomplish with a mix of bNAbs.
The study shows that three bNAbs must be combined to effectively restrict viral escape and forecast the ideal make-up of such a bNAb cocktail. The research findings demonstrate how genetic information can be used to predict treatment outcomes and develop new methods of pathogenic control while also providing a reasonable HIV therapy design.
According to the study, computational methods for choosing bNAb combinations based on viral genetics may lead to preventing viral escape and increase the efficacy of HIV treatment.
According to senior author Armita Nourmohammad, an assistant professor of physics at the University of Washington, “our study suggests that utilizing genetic data can help us build more effective HIV medicines.” Our strategy could be helpful in developing treatments for other fast-evolving pathogens such as the Hepatitis C virus, bacteria that are resistant to antibiotics, or cancer tumor cells.
A group of proteins known as antibodies is produced by the immune system to identify and combat infections. bNAbs are a particular subset of antibodies that are specific to HIV infections and can recognize various HIV strains.
Broadly neutralizing antibodies presents a promising new method for treating or perhaps even curing viral diseases like HIV. However, clinical studies treating HIV with a single bNAb have revealed that some viral strains may survive the treatment and cause a rebound of viruses in the blood. Using combinations of bNAbs may likely be a much more effective strategy, but it can be challenging to determine which combinations work best.
According to senior author Colin LaMont, a scientist at the Max Planck Institute for Dynamics and Self-Organization in Germany, for this study, the authors proposed utilizing a computational approach to forecast the efficiency of combination therapies (bNAb combinations) based on the HIV genotypes.
The scientists examined the genomes of HIV viruses obtained during a 10-year period from 11 HIV-positive patients who were not receiving treatment using high-throughput DNA sequencing. These data were used to forecast which virus strains could be able to avoid treatment with various bNAbs and determine whether evolving to avoid bNAbs had any negative effects on survival. The information was then used to predict viral rebounds in three actual trials using bNAbs using computational techniques. The team’s computational approach was then utilized to create a set of bNAbs that is least likely to let any virus escape.
They also learned why some bNAbs target populations with higher HIV prevalence than others. For instance, some bNAbs, such as one by the name of 10-1074, work better against a variety of virus populations because the modifications that allow viruses to escape from 10-1074 also decrease the likelihood that those viruses would survive. Due to the rarity of escape mutations, other bNAbs, such as PGT121, are more effective against virus populations with reduced genetic diversity. Overall, the findings indicated that PG9, PGT151, and VRC01 are the three bNAbs that work best together.
In this study, the scientists demonstrated that using PG9, PGT151, and VRC01 together lowers the risk of a viral comeback to less than 1%, according to LaMont. It accomplishes this by concentrating on three different areas of the virus’ outer, or protective, envelope.
According to Nourmohammad, combining bNAbs, which are given intravenously every few months, with currently available anti-retroviral treatments, which call for daily doses, could further increase the long-term success of HIV treatment.
Anti-retroviral therapy limits the genetic variety of the viral population and lowers the probability of the formation of bNAb escape variants by decreasing HIV’s capacity to proliferate and produce new variants. According to the authors, future research is required to verify any possible advantages of combining anti-retroviral therapy with broadly neutralizing antibodies.
Story Source: Colin LaMont, Jakub Otwinowski, Kanika Vanshylla, Henning Gruell, Florian Klein, Armita Nourmohammad (2022) Design of an optimal combination therapy with broadly neutralizing antibodies to suppress HIV-1 eLife 11:e76004 https://doi.org/10.7554/eLife.76004 https://www.washington.edu/news/2022/07/22/hiv-antibodies/
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.