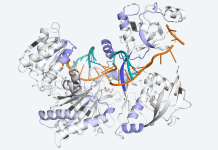
A significant complex of RAS-pathway proteins’ structure has been determined by scientists, revealing how known disease-causing mutations result in illness and offering prospective novel binding sites for cancer drugs.
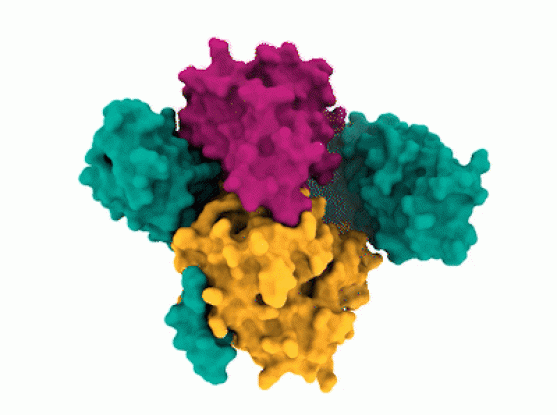
Image Source: https://www.broadinstitute.org/news/new-protein-complex-structure-reveals-possible-ways-target-key-cancer-pathway
One of the most well-known causes of cancer is RAS gene mutations, which result in tumor formation in about 25% of cancer patients. Broad Institute of MIT and Harvard researchers, in collaboration with the scientists from the Dana-Farber Cancer Institute, have determined the molecular structures of the RAS-pathway protein SHOC2 and two other proteins it binds. The three-protein SHOC2-MRAS-PP1C (“SMP”) complex regulates the RAS signaling system and aids in the survival of cancer cells that have RAS mutations.
The SHOC2-MRAS-PP1C holophosphatase complex serves as a crucial regulator of RTK-RAS signaling by removing an inhibitory phosphorylation event on the RAF family of proteins to enhance MAPK signaling. This integrated structure-function model of the RAS-pathway protein complex will guide the development of new drugs for cancer.
The high-resolution structure of the complex revealed using cryogenic electron microscopy, and X-ray crystallography provides hints as to how drugs can attach to it to block the RAS-pathway and halt the progression of cancer. The study offers a potential therapeutic strategy for a signaling pathway that has proven difficult to inhibit with drugs. The work was co-authored by researchers from Dana-Farber, the Broad’s Cancer Program, the Genetic Perturbation Platform, Deerfield Discovery and Development, a Deerfield Management Company subsidiary, the Center for the Development of Therapeutics, and the Broad’s Cancer Program (CDoT).
According to study co-senior author and Broad institute member William Hahn of the Cancer Program, by solving the structure, the researchers learned a lot about how SHOC2 acts in this circuit, which brings out areas in which you may therapeutically interfere. Hahn has several positions at Dana-Farber, including executive vice president, chief operational officer, and William Rosenberg Professor of Medicine.
Potential Therapeutic Target SHOC2
SHOC2 was first recognized as a potential therapeutic target in 2019 by Hahn, postdoctoral researcher Jason Kwon, and co-senior author Andrew Aguirre, an oncologist at Dana-Farber and a Broad associate member. Together with the other members of the Broad’s Cancer Dependency Map project, they looked at key genes in tumors with RAS protein mutations. At the time, cancer drugs known as MEK inhibitors were developed to focus on RAS protein mutations, but they were only successful in a tiny subset of cancer patients. The team made assumptions about whether an additional protein was enabling mutant RAS proteins to circumvent these inhibitors.
In order to find any potential genes that might be causing RAS-mutated cancer cells to become resistant to the effects of MEK inhibitors, the team used CRISPR-Cas9 to individually deactivate each gene throughout the entire genome in their 2019 work. When the SHOC2 gene was deleted, or its expression was decreased, it was discovered that the MEK inhibitors were very effective in killing cancer cells, proving that SHOC2 is the reason for resistance to MEK inhibitors.
SHOC2 – Horseshoe-Shaped Scaffolding Protein
In order to better understand SHOC2 as a prospective target, the team decided to solve its structural problems. At that point, Deerfield Discovery and Development scientists and CDoT joined the group.

Image Source: https://www.broadinstitute.org/news/new-protein-complex-structure-reveals-possible-ways-target-key-cancer-pathway
According to Kwon, the project manager and co-first author of the paper, the researchers identified SHOC2 genetically and understood that it was significant, but in order to therapeutically exploit it, the researchers needed to know the structure of the protein to find critical, functional sections.
Behnoush Hajian and Yuemin Bian, co-first authors and research scientists of CDoT, as well as Christopher Lemke, co-senior author, and director of protein science and structural biology, worked with the group. X-ray crystallography and cryogenic electron microscopy were used to establish the high-resolution structures of SHOC2 and the proteins with which it interacts.
The researchers observed that SHOC2 is a scaffolding protein with a smooth surface and a horseshoe structure, making it tough for drugs to bind to it. The full structural details of how SHOC2 assembles and binds to two crucial proteins, PP1C and MRAS, which are also a part of the RAS signaling pathway, were only revealed through further research. The researchers solved the structure of this three-protein SMP complex which identified new binding sites between the component proteins, targetting where researchers might be able to fit a drug.
According to Alex Burgin, senior director of CDoT and scientist at the Broad Institute, from the first look at the SHOC2 structure itself, it seemed that nothing could be done with it. “However, looking at the SMP complex, the researchers instantly noticed several binding pockets where drugs could bind. The complex structure fascinated the researchers as it opened up a new path ahead.
Elucidating Function from Structure
Together with colleagues from Broad’s Genetic Perturbation Platform, the researchers utilized a method called deep mutational scanning to assess the effects of mutations on each amino acid of SHOC2 on the protein’s functionality. Using the information they generated, they might be able to discover further links between SHOC2 mutations and other diseases. For instance, past studies have shown that mutations in this protein lead to Noonan-like Syndrome, a congenital condition that affects the development of physical traits.
The fact that every potential mutation has already been tried makes it feasible to quickly understand the structure, rationalize it, and be certain of the effects of those alterations, according to Hahn. This article indeed sheds light on how powerful that information is.
The researchers anticipate that their findings may assist medication producers in identifying substances that bind to the SMP complex to inhibit SHOC2 and clarify the clinical significance of SHOC2 mutations. To successfully counteract the effects of RAS mutations and stop the spread of cancer, it would be desirable to develop novel medications that target the SMP complex and could be used either alone or in conjunction with MEK inhibitors.
The researchers quickly progressed from the discovery of RAS-pathway protein SHOC2 as a high-priority target to a thorough structure-function roadmap for drug development because of excellent teamwork and the leveraging of various experts having knowledge in cancer cell signaling, functional genetic screening, and structural biology. According to Aguirre, the researchers expect that the study will help in developing much-needed novel RAS-mutant cancer drugs.
Story Sources: Kwon, J.J., Hajian, B., Bian, Y. et al. Structure–function analysis of the SHOC2–MRAS–PP1C holophosphatase complex. Nature (2022). https://doi.org/10.1038/s41586-022-04928-2
Sulahian, R et al. Synthetic Lethal Interaction of SHOC2 Depletion with MEK Inhibition in RAS-Driven Cancers. Cell Reports (2019). DOI:10.1016/j.celrep.2019.08.090
https://www.broadinstitute.org/news/new-protein-complex-structure-reveals-possible-ways-target-key-cancer-pathway
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.