Scientists from the Korea Advanced Institute of Science and Technology propose the PICASSO technique, which enables more than 15-color imaging of spatially overlapping proteins in a single imaging round without using any reference emission spectra. For a wide spectrum of researchers, the PICASSO technique offers a multiplexed imaging tool with great accessibility and accuracy.
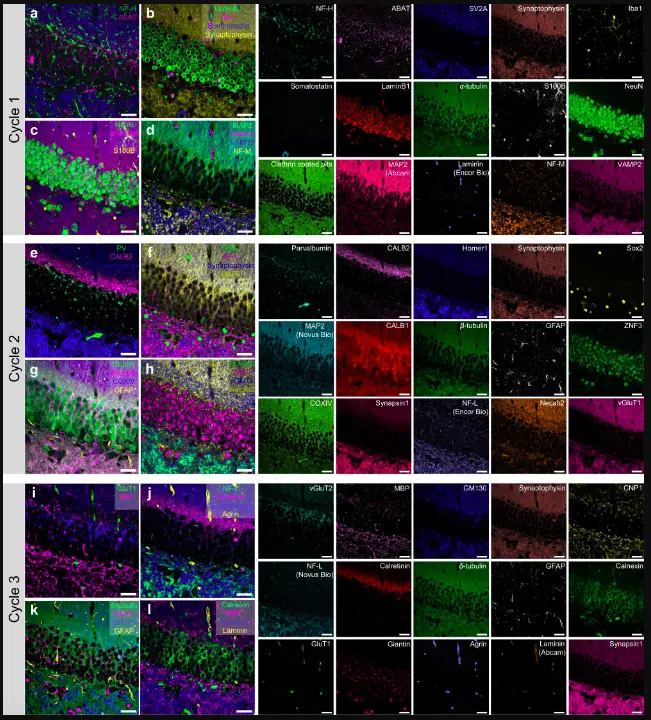
Image Source: PICASSO allows ultra-multiplexed fluorescence imaging of spatially overlapping proteins without reference spectra measurements.
According to the research published in Nature the PICASSO technique enables researchers to use more than 15 colors to scan and analyze our overlapping proteins by using artificial intelligence to clarify the spectrum color mixing of small chemicals used to label particular proteins and other objects of research interest. PICASSO can be used to attain strong multiplexing ability in different applications.
Pablo Picasso’s surreal cubist aesthetic transformed common characteristics into unrecognizable scenes, but a new imaging technique carrying his name may help us understand the most complex object of all: the brain.
When stimulated by light, fluorophores, the staining molecules, emit distinct colors; however, if more than four fluorophores are utilized, their released colors merge and overlap. By properly defining the matrix of mixed and unmixed images, researchers have already established strategies to eliminate this spectral overlap. This measurement relies on reference spectra, which are identified by locating clear photos of just one specimen stained with a fluorophore or of numerous, identically prepared specimens with just one fluorophore each.
According to the co-corresponding author Young-Gyu Yoon, professor in the School of Electrical Engineering at KAIST, due to the highly variable emission spectra of fluorophores depending on the subregions from which the spectra were recorded, such reference spectra measurement could be challenging to carry out in highly heterogeneous tissues, such as the brain. He explained that the approach would be ineffective and time-consuming because each of the subregions would require its own spectrum reference measurements. To solve this issue, the researchers created a method that does not rely on observations of reference spectra.
The technique, also known as PICASSO, is the “Process of ultra-multiplexed Imaging of biomolecules via the unmixing of the Signals of Spectrally Overlapping fluorophores.” Visualizing the several separate parts of a unit is referred to as ultra-multiplexed imaging. Each protein in a cell has a unique purpose, much like how each theatre plays a distinct film at a multiplex. Researchers can start to comprehend those roles by staining using fluorophores.
According to co-corresponding author Jae-Byum Chang, professor in the Department of Materials Science and Engineering at KAIST, the researchers developed a method based on information theory; unmixing is carried out by repeatedly decreasing the mutual information between mixed images. This facilitates precise information unmixing and lets us get away with the presumption that the spatial distribution of certain proteins is mutually exclusive.
The researchers used PICASSO to image a mouse brain in order to show off the tool’s capabilities. They carried out 15-color multiplexed imaging of a mouse brain with just one round of staining. Mouse brains, despite their tiny size, are intricate, multidimensional organs that might require a tremendous amount of resources to map. The researchers claim that PICASSO can expand the capabilities of existing imaging methods and enable the use of even more fluorophore shades.
In only three staining and imaging cycles, the team was able to image the mouse brain in 45 colors by combining one such imaging method with PICASSO, according to Yoon.
The multiplexed biomolecule imaging of cultured cells, tissue slices, and clinical specimens can be done with the use of PICASSO, according to Chang. The researchers believe that PICASSO will be helpful for a wide range of applications where spatial information about biomolecules is significant. Revealing the cellular heterogeneities of tumor microenvironments, particularly the heterogeneous populations of immune cells, which are intimately associated with cancer prognoses and the effectiveness of cancer therapies, is one use for the technology in this regard.
Story Source: Kim, G., Ahn, D., Kang, M. et al. Rapid species identification of pathogenic bacteria from a minute quantity exploiting three-dimensional quantitative phase imaging and artificial neural network. Light Sci Appl 11, 190 (2022). DOI: https://doi.org/10.1038/s41377-022-00881-x
https://news.kaist.ac.kr/newsen/html/news/?mode=V&mng_no=21511
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.






