Researchers from the University of Michigan have reported the discovery of a new plant protein fold that catalyzes the formation of macrocyclic peptides through a novel mechanism. Their findings provide insight into nature’s peptide cyclization strategies and expand the toolbox for bioengineering cyclic peptides as potential therapeutics.
Introduction
Cyclic peptides have gained increasing interest as a source of new drug leads. The connectivity between amino acid side chains confers enhanced stability and protein-binding capabilities compared to linear peptides. Hundreds of cyclic peptide natural products have been isolated from plants, fungi, and bacteria, exhibiting a wide range of bioactivities, but the biochemical mechanisms behind their biosynthesis have been largely unknown.
However, a major challenge in leveraging cyclic peptides as drugs is scaling their production. Only small amounts can be extracted from natural sources. While chemical synthesis is possible, the reactions are often low-yielding and not environmentally friendly. A better understanding of the biosynthetic enzymes producing cyclic peptides in nature could enable more sustainable and efficient biomanufacturing processes.
The researchers reported a new family of plant enzymes that naturally synthesize cyclic peptides through an unusual mechanism. Their findings provide a blueprint for engineering microbial systems to produce bioactive cyclic peptides.
RiPP Biosynthesis and BURP Proteins
Cyclic peptides synthesized by plants and microbes belong to a class of natural products called ribosomally synthesized and post-translationally modified peptides (RiPPs). RiPP biosynthesis involves extensive enzyme-catalyzed modifications of precursor peptides synthesized on ribosomes.
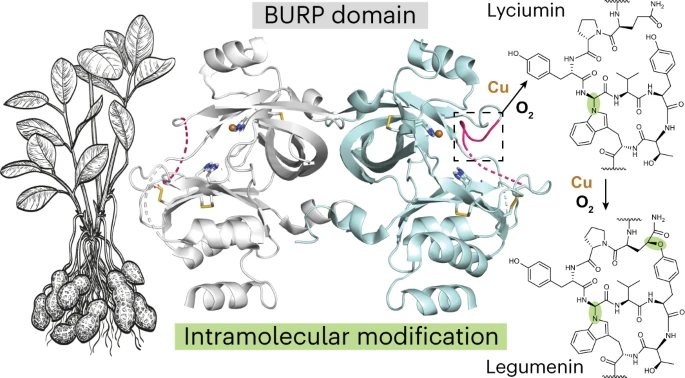
A subgroup of plant RiPPs contains macrocyclic modifications linking amino acid side chains within the peptide backbone. These macrocycles are installed by BURP domain-containing proteins, named after the four founding members: BNM2, USP, RD22, and PG1β.
BURP proteins consist of the defining BURP domain and short-conserved peptide motifs. They act as peptide ligases to macrocyclic specific motifs within their own precursor peptide sequences, liberating the cyclic peptide product via proteolytic cleavage.
Prior to this study, there was no structural or mechanistic information available on how BURP proteins catalyze macrocyclization. Elucidating their chemistry could inform engineering efforts to produce cyclic peptide drugs in microbes.
Discovering the Catalytic Mechanism of AhyBURP
The researchers focused their investigation on AhyBURP from the peanut plant. This USP-type BURP generates two macrocyclic peptides from its precursor peptide sequence: monocyclic lyciumin I and bicyclic legumenin.
Using X-ray crystallography, the researchers determined AhyBURP’s three-dimensional structure. The protein adopted a previously unobserved fold organized around two copper binding sites.
Enzyme assays revealed that AhyBURP catalyzes peptide macrocyclization using a mechanism requiring both dioxygen and radicals. The reaction proceeds sequentially, with the N-C(sp3) crosslink of lyciumin I forming first, followed by the O-C(sp3) linkage in legumenin.
This chemistry had not been observed for other known RiPP enzymes. AhyBURP was found to be capable of performing both the oxidation and final macrocyclization steps without additional enzymes.
Implications for RiPP Engineering
The researchers’ findings fundamentally expand the known repertoire of enzymatic strategies for RiPP biosynthesis. BURP proteins were shown to mediate intramolecular macrocyclization of core peptide motifs using a unique radical-mediated mechanism.
This chemistry enables the sequential installation of multiple macrocycles in a controlled one-pot reaction cascade. Such a process would be challenging to recreate efficiently via standard organic synthesis.
The structural insights into AhyBURP lay the groundwork for future protein engineering efforts. Its copper-binding residues and active site architecture could be modified to alter substrate scope and specificity.
Introducing BURP enzymes into microbial production strains is anticipated to provide more sustainable and scalable ways to access bioactive cyclic peptides. Their precursor peptides can also be engineered to generate analog libraries.
Outlook for Cyclic Peptide Drugs
Many cyclic peptides from plants and bacteria have promising therapeutic effects but are produced in miniscule amounts in nature. Unlocking their biosynthetic pathways now allows drug developers to explore their potential using synthetic biology techniques.
The legumenin cyclic peptide made by AhyBURP was previously shown to have anti-cancer cell properties. Elucidating its biogenesis may ultimately lead to a scalable production process.
The researchers also aim to discover and characterize other BURP enzymes that generate cyclic peptides with unique bioactivities. Combining these natural catalysts with synthetic biology and genome mining has the potential to expand our chemotherapeutic arsenal.
Conclusion
This research provided fundamental new insights into enzymatic strategy plants have evolved to construct pharmacologically relevant cyclic peptides. By elucidating the structure and mechanism of AhyBURP, the door has been opened to reengineering these natural peptide cyclases for biotechnological applications.
Understanding and harnessing nature’s biosynthetic machinery will enable more sustainable, efficient, and flexible production of promising cyclic peptide drug candidates. The discovery of this new plant biochemistry by the University of Michigan researchers represents an important step on that journey.
Article source: Reference Paper | Reference Article
Follow Us!
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.