In the vast and intricate realm of metagenomics, where microbial communities unveil a wealth of biomedical knowledge, a groundbreaking development has emerged. Scientists from Pennsylvania State University introduced MetagenomicKG, a new knowledge graph built to take the exploration of metagenomic data to unprecedented heights. This revolutionary resource has the potential to change how researchers address the complexity of microbial ecosystems and provides a comprehensive and interconnected framework tailored to the unique needs of this emerging field.
Unraveling the Intricacies of Microbial Communities
The large amount and diversity of genomic content in microbial communities make metagenomics an affluent area of biomedical knowledge. However, traversing these complex societies and their vast unknowns often depends on a variety of reference libraries, each with a specific analytical purpose. From the Genome Taxonomy Database (GTDB) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) to the Bacteria and Viruses Bioinformatics Resource Center (BV-BRC), these repositories are essential for the genetic and functional annotation of microbial communities.
Despite their valuable contributions, inconsistent nomenclature or identifiers between these repositories present challenges for effective integration, representation, and use. Enter the Knowledge Graph, a powerful solution that organizes biological entities and their relationships into a coherent network, revealing hidden patterns and enriching our biological understanding with deeper insights.
MetagenomicKG: A Tailored Solution for Metagenomics
While knowledge graphs have shown potential in various biomedical fields, their application in metagenomics has remained underexplored until now. MetagenomicKG emerges as a pioneering knowledge graph specifically tailored for metagenomic analysis, integrating taxonomic, functional, and pathogenesis-related information from widely used databases and further linking these with established biomedical knowledge graphs to expand biological connections.
A Comprehensive Graph Overview
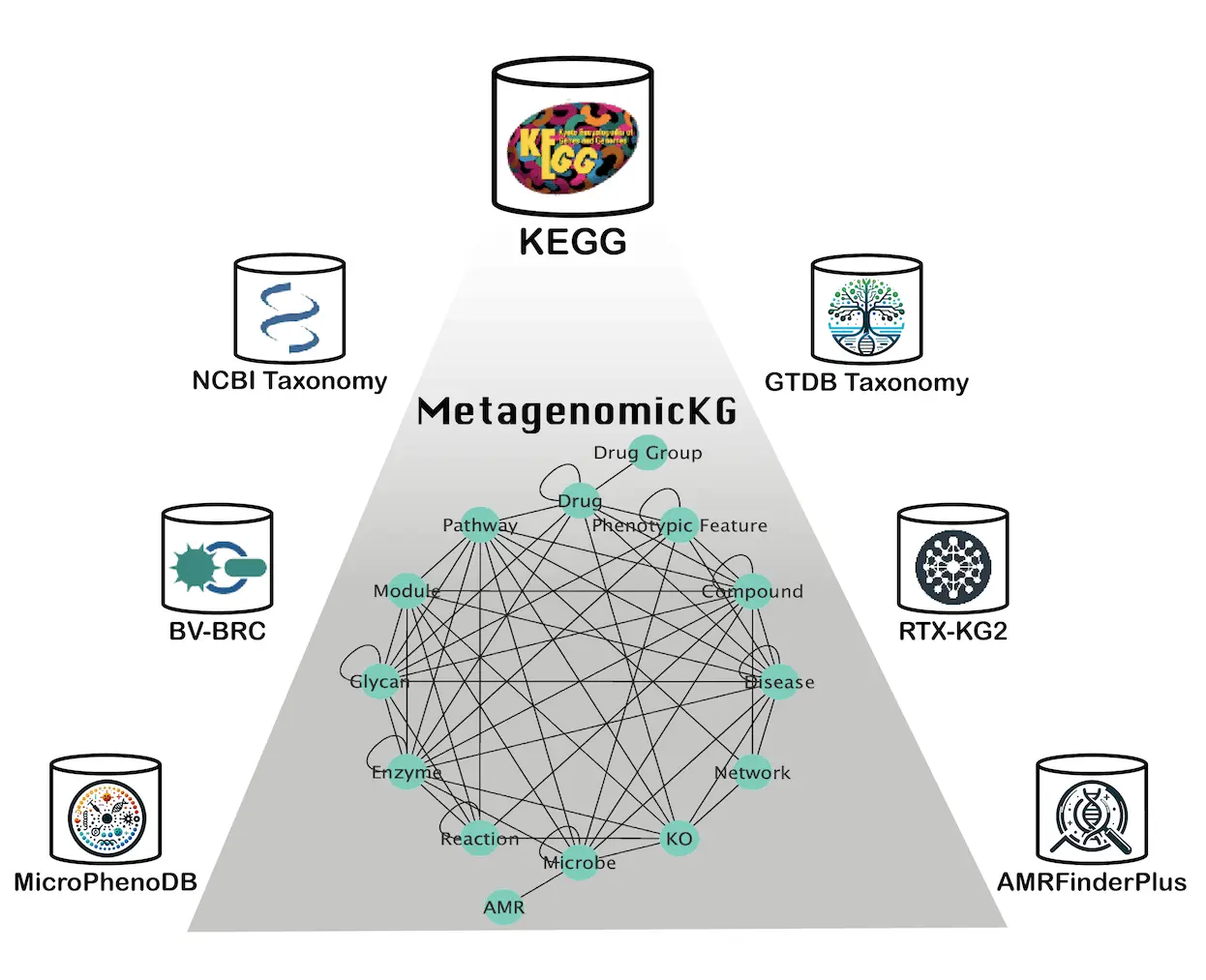
MetagenomicKG is a directed multigraph with 1.25 million nodes and 56 million edges connecting 14 types of nodes in accordance with the KEGG databases: Microbe, Phenotypic Feature, Disease, KEGG orthology (KO), Compound, Reaction, Drug, Glycan, Enzyme, Drug Group, Antimicrobial Resistance (AMR), Network, Pathway, Module.
To enable interoperability with other knowledge bases, MetagenomicKG adheres to the semantic schemas of existing biomedical knowledge graphs, mapping node types and edge types to the standard node category and predicates defined by the Biolink model – a comprehensive, open-source framework that aims to standardize and facilitate the interoperability of diverse biological and translational science data across biomedical knowledge graphs.
The MetagenomicKG’s construction process involves automated workflows using Python and shell scripting, with data stored in tabular format and imported into the Neo4j graph database. Identifiers and names of biological entities are harmonized using tools like GTDB-tk, KofamKOALA, and NCBI AMRFinderPlus Prediction and linked through ontology mapping and UMLS search. The graph’s extensibility allows for incorporating new data sources, reflecting the continuous evolution of metagenomic research.
Integrating Diverse Data Sources
At the heart of MetagenomicKG lies a meticulously curated integration of seven data sources, categorized into four major categories: Taxonomy, Functional Annotation, Pathogen Characterization, and Existing Biomedical Knowledge Graphs.
- Taxonomy: Incorporating the standardized, genome-based GTDB Taxonomy for bacteria and archaea and the NCBI Taxonomy for viral and fungal taxonomy information.
- Functional Annotation: Leveraging the comprehensive KEGG database, which stores molecular and functional annotations based on genome sequences and high-throughput data.
- Pathogen Characterization: Integrating the BV-BRC platform, providing abundant data for bacterial and viral pathogens, as well as the MicroPhenoDB database, mapping relationships between pathogenic microbes, their core genes, and human disease phenotypes.
- Existing Biomedical Knowledge Graphs: Harnessing the power of RTX-KG2, one of the largest open-source, Biolink model-based standardized Biomedical Knowledge Graphs, ensuring the integration of extensive knowledge from various databases.
Empowering Metagenomics Research: Use Cases
Several use cases demonstrate the promising utility of MetagenomicKG and its potential in advancing metagenomic research:
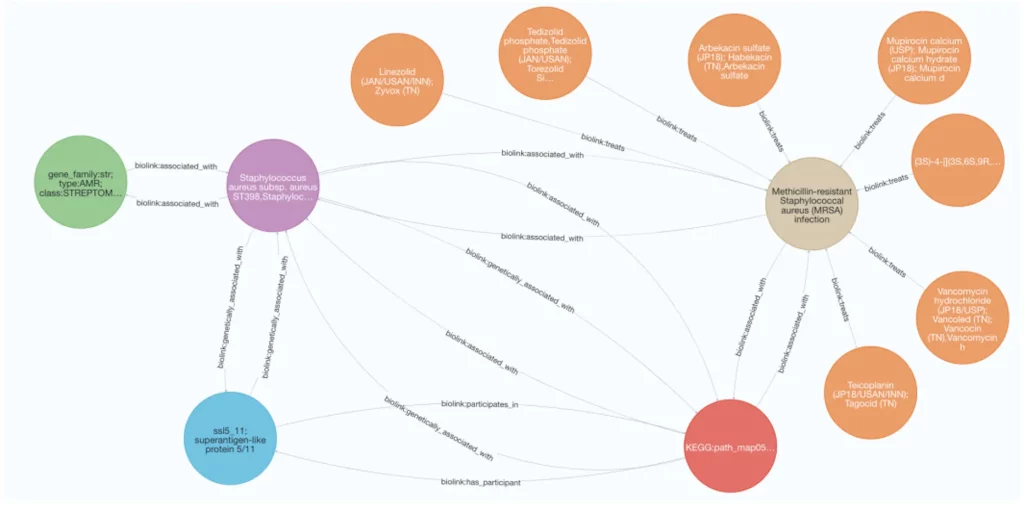
- Hypothesis Generation and Exploration: MetagenomicKG enables researchers to understand the complex relationships among pathogens, AMR-associated proteins, KEGG-based function annotations, diseases, and relevant drugs. It is a powerful tool for generating and evaluating biological hypotheses, fostering new insights into the intricate interplay between microbes and diseases.
- Sample-Specific Graph Embeddings: By embedding taxonomic profiling data with the topological structure of MetagenomicKG, researchers can derive sample-specific graph embeddings. These embeddings have demonstrated the ability to better distinguish metagenomic samples from different body sites compared to taxonomic profiling results alone, enhancing our understanding of microbial communities across diverse environments.
- Pathogen Identification: Combining graph neural network models with MetagenomicKG has shown improved accuracy in identifying pathogens, particularly for those pathogenic microbes with less sequence similarity to known pathogens. This innovative approach leverages the comprehensive knowledge within MetagenomicKG, advancing our capabilities in detecting and characterizing potential threats to human health.
Extensibility and Interoperability: A Future-Proof Foundation
MetagenomicKG is constructed from the fundamental and well-curated functional and molecular data of biological systems on the Kyoto Encyclopedia of Genes and Genomes (KEGG). By enriching KEGG with additional metagenomic data (taxonomy, antimicrobial resistance [AMR], and the associated diseases and drugs), MetagenomicKG provides researchers with a new innovative tool to elucidate the biological mechanisms of the microbiome on human health.
Furthermore, all nodes and edges in MetagenomicKG are annotated using the standardized ontologies of the Biolink Model, which enables easier extension and interoperability with other Biolink Model-based knowledge graphs deployed on SPOKE, KG-COVID19, BioThings Explorer, and mediKanren. This allows vast possibilities for collaborative research and the seamless integration of diverse knowledge sources.
Conclusion
MetagenomicKG is a benchmark in metagenomics and one of the first knowledge graphs tailored to the specific needs of metagenomics. The vast and heterogeneous data it provides, organized around rigorous ontologies and modeled via the newest developments in knowledge Graphs’ technology, will allow researchers to dive deeper into the complex interactions between microbial communities.
MetagenomicKG offers a stepping stone on the path to inform how knowledge graphs can support metagenomic knowledge creation and, in return, how the knowledge charts the global microbiome discovery landscape. Given its extensible focus and interoperability, MetagenomicKG can grow with and around its knowledge domain to be continually improved and extended by the metagenomic research community as a living knowledge graph.
Article source: Reference Paper | MetagenomicKG source code and technical details are available on Github | Neo4j instance for accessing and querying this graph is available at http://mkg.cse.psu.edu:7474.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.