Recent research has demonstrated that generative AI has the ability to greatly speed up drug development in the pharmaceutical industry. Currently, it can take several years for pharmaceutical companies to create drugs capable of addressing and potentially eradicating human diseases. To address this, scientists from The Ohio State University, USA, developed a generative framework G2Retro to substantially speed up the drug development process.
Understanding Retrosynthesis and the Need for Predictive Models
Retrosynthesis, the process of designing a synthesis route for a target molecule, has long been a fundamental challenge in organic chemistry. With the advent of artificial intelligence (AI), researchers have turned to machine learning algorithms to expedite the retrosynthetic planning process. One promising approach in this field is the utilization of graph generative models. In particular, the G2Retro model has emerged as a groundbreaking two-step graph generative model that demonstrates remarkable potential in retrosynthesis prediction. In this article, we will delve into the workings of G2Retro and explore how it is revolutionizing the field of organic synthesis.
Before we delve into the specifics of G2Retro, let’s understand the concept of retrosynthesis and the challenges associated with it. Retrosynthesis involves breaking down a target molecule into simpler precursors, ultimately identifying viable synthetic routes. This process traditionally relies on the intuition and experience of expert chemists, which can be time-consuming and subjective. Consequently, the development of predictive models that can automate retrosynthetic planning has garnered significant interest.
The Power of Graph-Generative Models
Graph generative models have gained prominence as a powerful tool for retrosynthesis prediction. These models represent molecules as graphs, with atoms as nodes and chemical bonds as edges. By learning from vast databases of known reactions, graph generative models can generate new molecules and predict viable retrosynthetic routes. The ability of these models to capture structural information and chemical transformations makes them ideal for solving complex retrosynthesis problems.
G2Retro: Advancing Retrosynthetic Analysis for Enhanced Drug Development
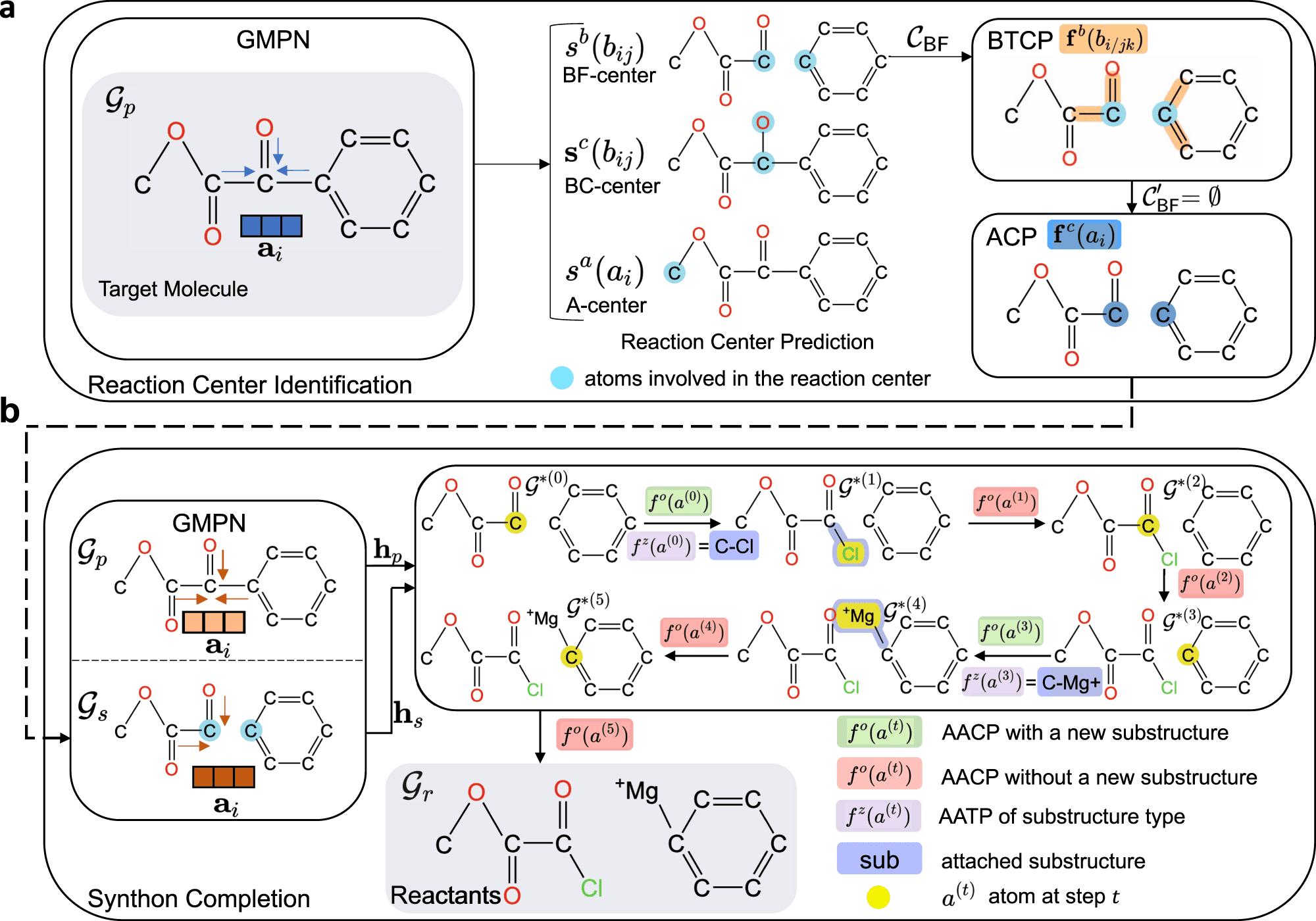
G2Retro, a state-of-the-art two-step graph generative model, has emerged as a breakthrough in retrosynthesis prediction. Unlike traditional graph generative models that generate molecules in a single step, G2Retro splits the process into two distinct steps: scaffold generation and side-chain generation. This unique approach allows G2Retro to exploit the modularity of chemical structures and effectively explore a vast chemical space.
In the first step, G2Retro generates the molecular scaffold, capturing the essential core structure of the target molecule. The model utilizes graph neural networks (GNNs) to learn the underlying patterns and relationships within the dataset, enabling it to generate diverse and chemically valid scaffolds.
In the second step, G2Retro expands upon the generated scaffold by generating side chains. This process involves predicting the attachment points and types of side chains, resulting in a comprehensive retrosynthetic analysis of the target molecule. By breaking the retrosynthesis problem into two steps, G2Retro overcomes the limitations of previous models and offers enhanced predictive capabilities.
Incorporating a dataset comprising 40,000 chemical reactions collected from 1976 to 2016, scientists successfully trained the G2Retro model. This framework utilizes deep neural networks to generate potential precursor structures, which can be employed in the synthesis of specific chemicals. By “learning” from graph-based representations of the molecules involved, G2Retro showcases remarkable capabilities in generating new reaction predictions. According to the researchers, the model’s generating prowess is so exceptional that when presented with a molecule, it can swiftly produce hundreds of novel reaction forecasts. G2Retro may provide a variety of synthesis alternatives and pathways, as well as a mechanism to score those possibilities for each molecule.
Advantages and Future Implications
G2Retro exhibits several advantages, positioning it as a formidable contender in retrosynthesis prediction. Firstly, by dividing the generation process into the scaffold and side-chain steps, G2Retro ensures greater interpretability and modularity. Chemists can easily comprehend and manipulate the generated molecules, facilitating more informed decision-making.
Furthermore, G2Retro demonstrates impressive performance in terms of both accuracy and diversity of generated retrosynthetic pathways. The model’s ability to explore the chemical space more effectively allows it to propose alternative synthesis routes, enabling researchers to consider multiple options.
G2Retro’s predictive capabilities were put to the test in a case study involving four drugs currently available in the market: Mitapivat, utilized in the treatment of hemolytic anemia; Tapinarof, prescribed for various skin conditions; Mavacamten, employed in systemic heart failure treatment; and Oteseconazole, used for combating female fungal infections. G2Retro successfully generated precise synthesis routes for these patented drugs while providing additional alternative routes that prove advantageous and feasible for synthetic purposes.
Looking ahead, G2Retro holds tremendous potential for further advancements in the field. The continuous integration of larger and more diverse reaction databases will enhance the model’s learning capacity. Additionally, fine-tuning the model to account for reaction conditions and stereochemistry will refine its predictions, making it an even more indispensable tool for chemists worldwide.
Conclusion
The G2Retro model, with its innovative two-step graph generative approach, has emerged as a game-changer in the realm of retrosynthesis prediction. By breaking down the synthesis planning process into the scaffold and side-chain generation, G2Retro offers improved interpretability, modularity, and predictive capabilities. With this discovery, retrosynthetic planning might be revolutionized, becoming quicker, more effective, and less dependent on human intuition. It is fascinating to see how machine learning and artificial intelligence are changing the discipline of organic chemistry. G2Retro serves as a prime example of how cutting-edge technologies can complement human expertise, opening up new avenues for discovery and innovation in the synthesis of complex molecules. With ongoing research and refinement, G2Retro is set to empower chemists globally and accelerate the progress of organic synthesis.
Article Source: Reference Paper | Reference Article | G2Retro: Web Portal
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.