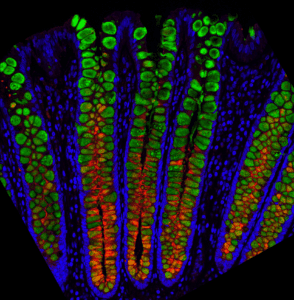
Scientists from the University of North Carolina School of Medicine, led by Scott Magness, Ph.D., sequenced the genes expressed in single cells from human digestive tracts to discover new cell-type characteristics and learn more about essential cell functions like nutrient absorption and immune defense.
You could feel uneasy in your tummy if you get nervous. If you consume chili, your stomach may revolt, yet your friend can eat anything and feel fantastic. Ibuprofen may be taken like candy with no side effects, but your friend’s stomach may bleed, and she may not receive pain relief. What is the reason behind this? The short answer is that we’re all unique. The following questions are: how different are they actually, and what does this signify for health and disease? Answering questions is far more challenging, but Scott Magness, associate professor and senior author of the paper, offers some intriguing scientific solutions.
The Magness group employed entire human gastrointestinal (GI) tracts from three organ donors for the first time to demonstrate how cell types differ throughout all parts of the intestines, sheding light on cellular activities, and highlight gene expression variances between these cells and across individuals.
The study, which was published in Cellular and Molecular Gastroenterology and Hepatology, paves the way for researchers to investigate the numerous components of gut health in far greater detail and resolution than previously done.
Image Source: A proximal-to-distal survey of healthy adult human small intestine and colon epithelium by single-cell transcriptomics.
The researchers demonstrated that understanding each cell type’s role in crucial processes including nutrition absorption, parasite defence, and the creation of mucus and hormones that influence eating behaviour and gut motility is conceivable. They also discovered how receptors and sensors in the gut lining interact with the environment, as well as how medications interact with various cell types.
The Gut’s Sensitivity
Consider a typical pharmaceutical ad voiceover, in which the voice actor cheerfully recites probable side effects, including diarrhea, vomiting, intestinal bleeding, and other unpleasant collateral damage. The Magness group is trying to figure out why such side effects occur, right down to individual cells, their roles, locations, and DNA.
The epithelium, a single-cell thick layer that separates the lining of the intestines and colon from everything else, was the focus of the Magness lab’s study. The epithelium, like other cell types and the microbiome, is vital to human health, and scientists have been studying it for years. Researchers could only collect minuscule biopsies the size of rice grains from a few areas of the digestive system up until now, mainly the colon or a few small intestine portions.
“Such exploration would be like looking at the United States from space but only investigating what’s going on in Massachusetts, Oklahoma, and California,” Magness said. “To really learn about the country, we’d want to see everything.”
“Not only do we want to identify where the cells are located, but we want to know exactly which cell types do what, and why,” Burclaff said. “So, staying with the map analogy, we don’t want to just say, ‘oh, there’s North Carolina’. We want to know where to get the best barbecue. We want a ground level view to know as much as possible.”
Researchers mash-up tiny rice-sized samples to identify all epithelial cell types and understand some of their general characteristics. Magness’ method involved collecting hundreds of individual cells from every portion of the lower digestive tract (small intestine and colon) to form an atlas and then studying the potential roles of these cells using the genes expressed by each cell. Knowing all of this would add to our understanding of the gut epithelium and, perhaps, stimulate future researchers to investigate each cell’s role in biology, illness, and the unpleasant scenario of medication side effects.
Magness required two things to perform such a deep individual cell dive: improved technology and human digestive systems in their entirety.
Biological Data for Research
The state-of-the-art RNA sequencing technology was acquired by UNC-Chapel Hill several years ago to create the Advanced Analytics Core Facility via. the UNC Center for Gastrointestinal Disease and Biology, that contributed to the development the scientific and intellectual heft.
The Magness Group obtained human digestive tracts in a study partnership with HonorBridge’s organ donation services. HonorBridge staff interacts with the Magness Group to donate transplant-grade organs for research when intestines are removed for transplant and are not claimed by higher-priority groups.
Image Source: A proximal-to-distal survey of healthy adult human small intestine and colon epithelium by single-cell transcriptomics.
The Magness lab gets complete digestive tracts, each around 15 to 30 feet long, six to eight hours after harvest. They remove the epithelial layer, which, while being only one cell thick, is one continuous linked piece of tissue. The epithelium is then broken down into individual cells with the help of enzymes. They did the same thing with organs from three other donors for this investigation.
The Magness group uses sequencing technology to evaluate gene expression by first extracting RNA from each cell at the same time keeping each cell separate and then running single-cell sequencing, which takes a snapshot of which genes and how much each intestinal cell is expressing.
“The picture we get from each cell is a mosaic of all the different types of genes the cells make, and this complement of genes creates a ‘signature’ to tell us what kind of cell it is and potentially what it is doing,” Magness said. “Is it a stem cell or a mucous cell or a hormone-producing cell or an immune-signaling cell?
Joseph Burclaff, postdoctoral fellow and co-first author of the paper expressed that, “We were able to see the differences in cell types throughout the entire digestive tracts, and we can see different gene expression levels in the same cell types from three different people. We can see the different sets of genes turned on or off in individual cells. This is how, for instance, we might begin to understand why some people form toxicity to certain foods or drugs and some people don’t.”
The massive volume of data generated is a crucial issue with this type of research. In just one cell, the single-cell sequencing picks up around 11,000’reads,’ or individual samples of gene products, and in many thousands of distinct cells, each with various combinations of the 20,000 or so human genes that are switched on or off. For all 12,590 cells in the research, this results in about 140,000,000 data points that must be converted into a “visual” format so that scientists can make sense of the massive amount of data.
“The human brain can only comprehend two dimensions, three is challenging,” tells Magness. “Add time, and it’s even trickier to comprehend what a single cell is up to. The amount of data our experiments produced was basically millions of dimensions all at once.”
Jarrett Bliton, a graduate student and co-first author of the paper created computational algorithms to filter the data, resulting in a manageable data set that includes cell populations from all parts of the gastrointestinal system. Bliton could then computationally identify each cell type from each location based on what Magness and other researchers had already learned about each cell type. The data was then plotted in a way that humans could comprehend and analyze.
The scientists learned a lot about each cell type after taming the massive data. Consider the tuft cell, which was found 40 years ago and is named for the appearance of tufts of hair on its surface. These tuft cells, express genes that are identical to those found in taste buds on the tongue. Other researchers revealed that these tuft cells detected worm infestations and signaled the immune system to go on the offensive. The Magness team discovered that tuft cells had a collection of genes that are considered critical for perceiving and “tasting” different types of intestinal material so that the immune system may be notified if necessary.
“Not only did we describe every single cell type and every single gene they express individually, but we also looked at potential functions,” Burclaff said. “If you look at intestinal mucus, which is a complex mixture that protects the cells, we show which cells express various mucin proteins, how much, and in which regions of the digestive tract. We looked at where specific enzymes that digest food are expressed. We looked at cells with anti-inflammatory gene expression and synapse genes where the gut is probably connected to nerves so it can talk to the rest of the body. We looked at aquaporins, proteins involved in transferring water through the intestinal membrane.”
The Magness team discovered a whole new degree of variety in possible functions that had previously gone unnoticed due to the mash-up of biopsy samples.
All epithelial receptors — cell surface proteins that communicate with other cells and chemicals, as well as the gut environment – were investigated. Magness and colleagues were able to observe which receptors were expressed the most and in which cell types, providing fresh insight into how cells interact with gut contents, including nutrients, bacteria, toxins, and medications.
“As far as we know, we’re the first to do this kind of analysis across the length of the human gut from three full donors,” Bliton said. “We can look at each cell type and predict which pharmaceuticals might affect which cell type individually.”
For example, there is a class of medications that are used to treat inflammatory bowel disease. These treatments are designed to target specific immune cells that cause inflammation. However, the Magness group discovered that specific epithelial cells express the same genes as the immune cells that would be targeted. This research suggests that epithelial cells may have unintended “off-target” actions that might cause adverse effects.
“This was not known,” Burclaff said. “Lots of drugs have bad GI side effects. And it could be because the drugs are affecting individual cells along the entire length of the GI tract. We show where these receptors are most expressed and in which cell types.”
This is only one of the Magness lab’s early research findings.
“We want the scientific, medical, and pharmaceutical community to use what we’ve found,” Magness said. “We adopted an analytic approach to methodically address each cell type, produce easy-to-read and accessible spreadsheets for most scientists, and show several examples of what we can be discovered with this kind of high resolution, precision approach.”
Story Source: Burclaff, J., Bliton, R. J., Breau, K. A., Ok, M. T., Gomez-Martinez, I., Burclaff J, Bliton RJ, Breau KA, Ok MT, Gomez-Martinez I, Ranek JS, Bhatt AP, Purvis JE, Woosley JT, Magness ST. (2022) A proximal-to-distal survey of healthy adult human small intestine and colon epithelium by single-cell transcriptomics. Cell Mol Gastroenterol Hepatol .https://news.unchealthcare.org/2022/02/scientists-map-entire-human-gut-at-single-cell-resolution/
Background Image Description: Protein MUC5b (red) in crypt-resident goblet cells; MUC2 (green) in all goblet cells.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.