Harvard University researchers introduce RNA3DB, a dataset crafted from the Protein Data Bank (PDB), addressing challenges in RNA structure prediction. In response to limitations in existing literature and recent findings in CASP15, RNA3DB offers a non-redundant dataset for training and benchmarking deep learning models.
With the success of deep learning in protein structure prediction, there has been growing interest in applying similar techniques to RNA structure prediction. However, RNA introduces unique challenges due to the sparser availability and diversity of experimentally determined RNA structures compared to proteins. Recent assessments have shown deep learning models underperform traditional methods for RNA structure prediction. In this article, we’ll discuss the challenges facing deep learning for RNA structure prediction and a new dataset called RNA3DB that aims to address these issues.
The Promise and Pitfalls of Deep Learning for RNA
After DeepMind’s AlphaFold dramatically improved protein structure prediction, many researchers quickly jumped on board to adapt these lessons to RNA structure prediction. Both involve predicting 3D structures from 1D sequences that are strongly dependent on the sequence. So, it seemed promising to apply deep learning techniques successfully for proteins to RNA as well.
Several papers reported impressive results for predicting both secondary and tertiary RNA structures. However, RNA researchers have not considered deep learning to be state-of-the-art for structure prediction. Issues arose regarding generalization to unseen sequences, with models overfitting on training data. This behavior was already known for probabilistic models trained on RNA structures since 2012.
More concerning, at the recent CASP15 structure prediction assessment, deep learning models were outperformed by traditional methods for the RNA-only targets. Since then, more publications have tried applying deep learning to RNA, often with inflated performance due to overlaps between training and testing data.
Unique Obstacles for Deep Learning with RNA
So why has deep learning for RNA structure lagged behind proteins? While some issues may be addressable with engineering solutions, the limited quantity and diversity of RNA structures present fundamental obstacles.
The number of available proteins in the PDB far exceeds that of RNAs. After filtering, the PDB contains 70 times more protein chains than RNA! With deep learning so dependent on data quantity, this scarcity of RNA structures poses a major challenge. And while PDB RNA entries are increasing yearly, they remain dwarfed by proteins.
The PDB also lacks optimization as a dataset for deep learning. Many RNAs are fragments rather than full structures, with extensive repetition of the same sequences. Specialized datasets are needed to cluster non-redundant RNAs and divide training, validation, and testing.
Other factors also contribute to the difficulty of RNA structure prediction compared to proteins. RNA has a more complex backbone geometry and global interdependence between secondary and tertiary structures. However, the limited data seems to be the most pressing issue holding back deep learning for RNA.
Introducing RNA3DB: A Dataset for Deep Learning on RNA
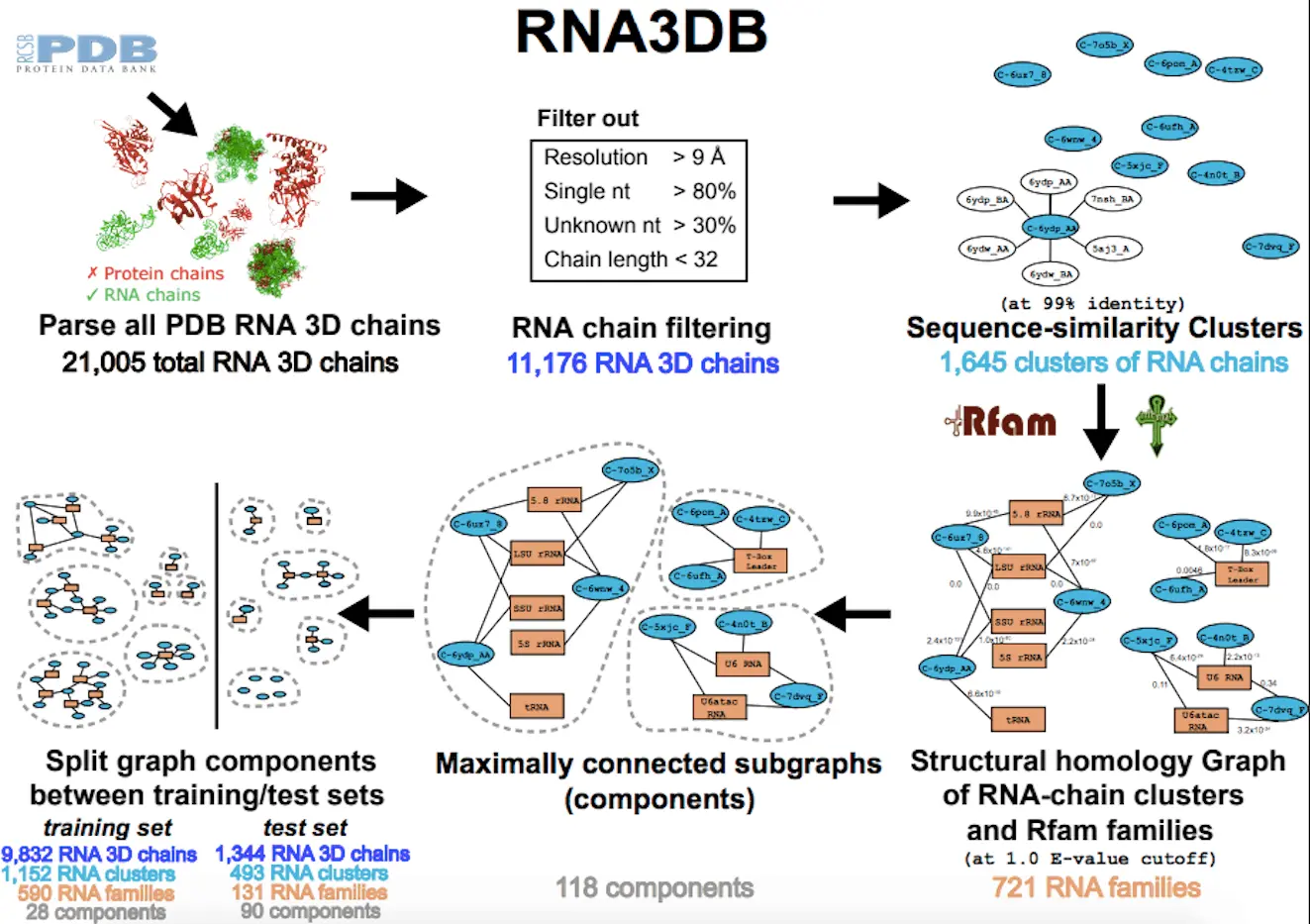
To address these data challenges, researchers have developed RNA3DB, a new dataset based on PDB RNA structures tailored for deep learning. RNA3DB clusters RNA chains into distinct, non-redundant groups by both sequence and structure. This enables proper division into training, validation, and test sets.
RNA3DB contains 21,005 RNA chains from the PDB as of early 2024. After filtering for length, resolution, sequence complexity, and redundancy, 11,176 chains remained. These grouped into 1,645 clusters sharing 99% sequence identity.
Adding Rfam family classification created a graph with RNA chain clusters, Rfam nodes, and edges indicating membership. The final RNA3DB contains 118 non-redundant components, with the largest containing 935 RNA chain clusters across 119 Rfam families.
This comprehensive classification provides maximally distinct RNA structures for a rigorous investigation into generalization. The creators hope RNA3DB will become a standard for training and benchmarking deep learning approaches to RNA structure prediction.
RNA3DB Analysis Reveals Obstacles for Deep Learning
Analysis of RNA3DB exposes just how limited the RNA structural data is compared to proteins. About 1 in 10 RNA PDB chains prove redundant or too short for deep learning methods.
The largest cluster contains 629 chains from the Thermus thermophilus ribosome. Half the components contain just a single RNA family and cluster. This highlights the diversity challenge – the few distinct RNA structures known make generalization difficult.
The median RNA cluster size is just 2. The median Rfam family node has edges to 2 RNA clusters. Contrast this with image datasets used in deep learning with orders of magnitude more examples.
So, while deep learning thrives on huge quantities of data, the PDB provides limited RNA data sparseness. This scarcity alone could explain poor deep learning performance for RNA structure prediction thus far.
The Potential of RNA3DB for Advancing Deep Learning
By exposing the data challenges facing deep learning for RNA, RNA3DB enables the proper assessment of model performance. The non-redundant clustering ensures rigorous, realistic testing without leakage between training and test sets.
The dataset creators are hopeful RNA3DB can reveal whether deep learning can generalize from extremely sparse data to unseen RNA structures. Some findings in image analysis suggest deep learning can be generalized despite small datasets.
With customizable filtering and splitting, RNA3DB supports investigating performance in different low-data regimes. This rigorous benchmarking is essential to determine if deep learning can overcome RNA’s data limitations or if hybrid approaches remain preferable.
Either way, RNA3DB provides a vital, reusable tool for standardizing the development and evaluation of deep learning for RNA structure prediction. Shared benchmarks lower barriers to progress. The dataset helps the field transparently assess the capabilities and limitations of different techniques.
Conclusion
RNA structure prediction remains crucial for deciphering the enormous regulatory roles of RNA in cells. While deep learning has accelerated protein structure prediction, unique obstacles have slowed similar progress for RNA. Data quantity and diversity represent fundamental challenges that are not easily surmounted. RNA3DB aims to enable rigorous focus on these issues to determine if they intrinsically limit deep learning for RNA.
Suppose deep learning can eventually generalize from extremely sparse data, datasets like RNA3DB to drive progress. However, hybrid approaches combining learned and physics-based models may continue to outperform pure deep learning.
Either way, RNA3DB provides an important new resource to strengthen training, evaluation, and transparency. Shared benchmarks and evaluations will help unlock the potential of both deep learning and complementary techniques for tackling the RNA structure prediction challenge.
Story source: Reference Paper | The database and code for RNA3DB are available on GitHub
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.















[…] Revolutionize RNA Structure Prediction with RNA3DB: The Pinnacle Dataset for Training Deep Learning … […]