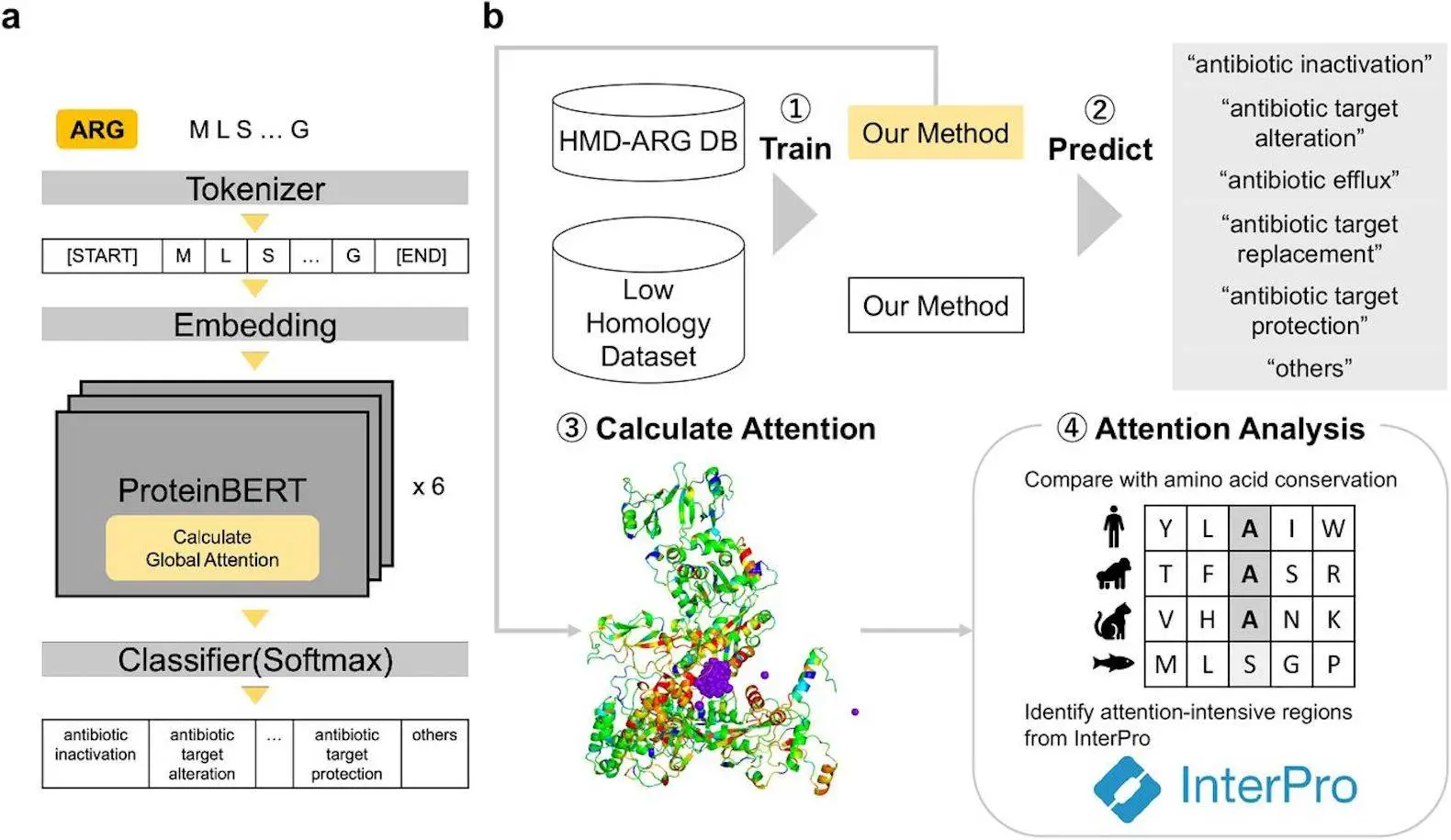
Scientists from Waseda University and Hitachi, Ltd., Japan, developed a novel approach for predicting antibiotic resistance genes (ARGs) resistance mechanisms using Protein-BERT, a protein language model based on deep learning. This model excels at predicting resistance for genes with low similarity to known sequences. By analyzing biologically relevant features like conserved residues and binding sites, Protein-BERT offers valuable insights and interpretability for tackling antibiotic resistance.
Antibiotics, which used to astonish physicians, have become less potent. Bacteria quickly develop immunity, thus posing a significant threat to global health. By 2050, according to WHO, antibiotic-resistant bacteria may cause up to 10 million deaths every year.
What are ARGs, and Why Do They Matter?
Think of bacteria as tiny fortresses, complete with specific entry gates and internal functions that allow them to receive only necessary things for their bodies and keep out poisonous substances. Antibiotics target these gates, while others interfere with inner mechanisms. However, bacteria can obtain mutations in their genes, which is akin to the creation of new gates or modifying existing ones, leading to rendering antibiotics useless. These mutated genes are called ARGs.
The problem is that the number of ARGs has become so large, and the number of various types is constantly increasing. Traditional methods to identify resistance mechanisms look at comparing ARGs with known sequences. It works well for well-studied genes but fails when analyzing completely new genes.
The Rise of Protein Language Models
Protein language models are leading the way in artificial intelligence as the field evolves at a breakneck speed. PLMs are built from huge datasets of protein sequences and their functions, which helps them find patterns and connections between structure and function.
Yagimoto et al. (2024) tapped into the potential of Protein-BERT, an example of a PLM to address ARGs. There are several advantages to their strategy:
- A New Genes’ Accuracy: In contrast with conventional methods, Protein-BERT can investigate ARGs that have little similarity with known genes; hence, it can adapt to resistance that changes regularly.
- Interpretability: Unlike other interpretations, Protein-BERT will not only predict the mechanism of resistance but also where in that protein sequence such resistance happens. It is like a window through which we can see how bacteria become resistant.
How Does it Operate?
ProteinBERT works like a well-trained detective.
Imagine that at a crime scene, you feed Protein-BERT, the protein sequence of an ARG. The sequence is analyzed by looking for patterns and suspicious characters (amino acids) that could be involved in the “crime” of antibiotic resistance.
Here’s How It Works:
- Training: Protein-BERT is trained on a huge dataset of known ARGs with resistance mechanisms. This enables it to decipher protein lingua franca and how diverse sequences may refer to various kinds of resistance.
- Prediction: Whenever given an additional ARG sequence, Protein-BERT scrutinizes its contents in search of any similarities or patterns that are discovered within the training data resulting from this. From these comparisons, the new arginine biosynthesis pathway predicts the most likely resistance mechanism.
- Attention Analysis: Now, this is where Protein-BERT shines. It’s not just about giving you an answer; there is a why behind that answer. By analyzing those components in protein sequence that are attended more by Protein-BERT (called attention), scientists can understand what specific amino acid(s) or regions are important for predicted resistance mechanism.
A Peep into the Future
We are now able to predict and understand resistance mechanisms accurately using ProteinBERT, which is potentially a powerful tool in the fight against antibiotic resistance.
Some Possible Developments Include
New antibiotic design: Scientists can develop drugs with weaknesses in certain areas once they understand resistance.
Treatment optimization: Doctors will get to know what causes drug resistance in individual cases, hence administering appropriate antibiotics.
Monitoring resistance evolution: In this regard, ProteinBERT keeps track of changes in genes leading to resistance over time.
Join the Struggle!
Antibiotic resistance is a frightening aspect of our fight against infectious diseases; it is difficult and demands diverse solutions. The coming together of technology and partnership will not only enhance the effectiveness of antibiotics but also ensure their use in fighting infections for many years to come. While this new strategy by Yagimoto et al. may be seen as a breakthrough, it should never overshadow the responsible use of antibiotics or efforts toward alternatives via new antibiotic investigations.
Also, citizens and scientists can play a role in this anti-microbial struggle. To achieve this, we all have roles to play, such as practicing good hygiene, avoiding unnecessary use of antibiotics, and supporting research programs.
Do you have any questions about antibiotic resistance? Feel free to leave your responses below!
Article Source: Reference Paper | The source code is available freely on GitHub. The output results of the model are published at https://waseda.box.com/v/ARG-BERT-suppl.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.