Peptides, which are short chains of amino acids, have become increasingly important in the world of medicine and biotechnology. Their unique properties make them ideal candidates for applications ranging from hormone therapies to antimicrobial agents. However, designing new, functional peptides has always been a challenge, primarily because traditional machine learning models require enormous datasets, often thousands of examples, to accurately predict which sequences will work. This requirement has made advanced peptide discovery inaccessible to many smaller labs and academic groups that simply don’t have the resources to generate such large datasets.
The MDMI Platform: Smart Insights from Minimal Data
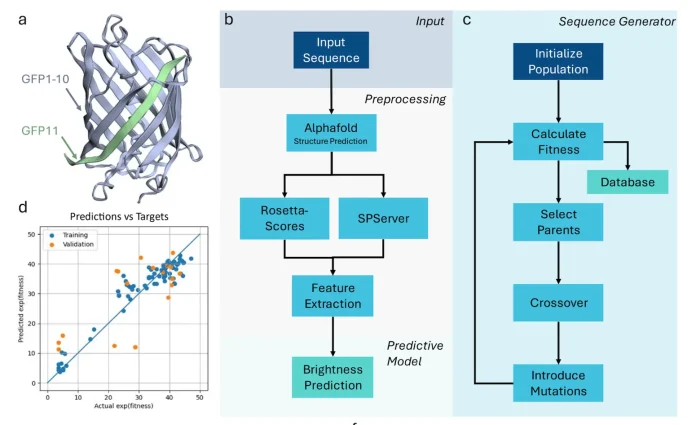
Recognizing this challenge, a group from the University of Toronto developed a unique platform called ‘Minimal Data Maximal Insight’ (MDMI). This system aims to maximize information retrieval from data as small as 100 peptide sequences. Rather than MDMI’s strategy of gathering data heavily overperforming computational modeling, smart experimental designs make each data point count. The platform has AlphaFold Multimer, which predicts the 3D structures of peptide-protein complexes. This offers more insight into the interactions between molecules. These structural predictions undergo evaluation through both statistical and physics-based scoring, assuring that only the most reliable candidates go further.
Evolutionary Algorithms: Mimicking Nature for Better Peptides
MDMI utilizes a genetic algorithm that emulates the process of natural selection to create new peptide sequences. It iteratively combines and mutates the best-performing sequences to traverse a peptide’s possible landscape while ensuring they meet essential criteria such as solubility and stability. This method allows researchers to target experimental work towards the most promising candidates, which significantly cuts down the time and expenses associated with peptide discovery.
Case Study: GFP11 and the Power of MDMI
The split Green Fluorescent Protein (GFP) system demonstrated the power of MDMI. In this model, a small peptide known as GFP11 associates with a larger fragment, GFP1-10, and they form a fluorescent protein together. This configuration allows a straightforward functional assessment of the peptide, which makes it a perfect test case for the platform. The team simulated the structures of over 1,300 different variants of GFP11 and executed their scoring system to project the most successful candidates. Then, they employed the genetic algorithm to create new sequences and settled on 40 frontrunners after laboratory testing.
Diversity and Functionality
The outcome was remarkable. More than sixty percent of the designed peptides exhibited sustaining more than twenty percent of the activity of the original GFP11, and almost a third surpassed sixty percent of wild-type activity. Even more astonishing, some of the best peptide sequences changed up to sixty-two percent of their amino acids and still performed well. This change dramatically increases the number of candidates that can be created without evoking an immune response or denaturing in therapeutic contexts.
What makes MDMI truly revolutionary is how easy it is to access. It transforms the use of open-source datasets into participant-led peptide discovery by employing computational tools and requiring only small datasets. Labs that previously didn’t have the means, whether time or money, to participate in traditional methods can now join the emerging field of peptide engineering. This approach isn’t just targeted at GFP; it can also be applied to other peptide-protein systems, including those pertinent to antimicrobial discovery and targeted drug delivery.
Conclusion
MDMI is a step forward for the entire discipline of computational biologics. The integration of structure-based modeling, machine learning, and evolutionary algorithms allows scientists to create functional and diverse peptides with little to no experimentation. It could expedite the creation of new treatments and broaden the reach of sophisticated peptide design to many more scientists.
Article Source: Reference Paper
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: OpenReview releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.