In living organisms, different types of proteins are essential for a variety of critical processes. The structural and functional diversity of many protein types is greatly influenced by post-translational modifications (PTMs), which allow cells to respond to changes in their environment and control intricate biological processes. Though they do not take into consideration the effects of PTMs, large protein language models (pLMs) such as ESM-2 and ProtT5 precisely capture the structural, functional, and physicochemical features of input protein sequences, they only scratch the surface of proteome diversity. By adding PTM tokens to the pLM training regimen, researchers bridge this significant gap in protein sequence modeling.
In this study, Duke University researchers introduce PTM-Mamba, a PTM-aware Protein Language Model that makes use of an extensive collection of PTM tokens to enhance performance on a range of PTM-specific tasks. The model vocabulary gets around Transformers’ quadratic time complexity using effective hardware-aware primitives. The first and only pLM that inputs and representations wild-type and PTM sequences in a unique way is PTM-Mamba. This makes it ideal for downstream modeling and design applications that are tailored to post-translationally modified proteins.
Introduction
The human genome contains more than 20,000 different proteins, making proteins the primary molecular machinery of nature. These proteins conduct a variety of biochemical activities, give cells vital scaffolding support, and move essential chemicals across organs and tissues. The functional capacities of eukaryotic proteomes are greatly increased by post-translational modifications (PTMs), including phosphorylation, acetylation, ubiquitination, and glycosylation. Crucial biomolecular functions include modulation of specific signaling cascades, cell division, DNA repair, localization dynamics, turnover of proteins, core enzyme activities, and protein-protein interactions are all impacted by PTMs. Several disease phenotypes, including cancer, neurodegeneration, and aging, are brought on by dysregulation of PTM installation. Capturing the distinctive characteristics of post-translationally changed protein sequences is, therefore, essential.
Protein sequences are a prime candidate for language modeling due to their hierarchical semantics and sequential structure. Latent embeddings that accurately encode pertinent physicochemical and functional information are produced using protein language models (pLMs), such as ProtT5 and ESM-2, which have been pre-trained on over 200 million genuine protein sequences. Numerous protein sequences with confirmed functional capabilities have been produced using autoregressive pLMs, such as ProGen and ProtGPT2. Protein sequence embeddings are most useful for precise downstream protein tertiary structure prediction, as exemplified by models such as ESMFold and AlphaFold2. PTM effects cannot be modeled in sequence or structural space, nevertheless, because none of these attention-based models takes PTM residues into account.
Understanding PTM-Mamba
Researchers trained the first pLM to explicitly incorporate PTM information, therefore addressing a fundamental gap in protein sequence modeling. To get around the quadratic temporal complexity of the traditional attention mechanism, the PTM-Mamba model, a recent breakthrough in state space modeling (SSMs), makes use of hardware-aware primitives. Training is done on a large-scale post-translationally changed protein sequence dataset, and the embeddings of PTM-Mamba and ESM-2 are dynamically combined in a gated method via a fusion module. As a consequence, performance on downstream PTM-specific supervised training tasks is improved. The results demonstrate that infusing PTM information into expressive, pre-trained pLM latent spaces separates latent representations of PTM sequences from their wild-type counterparts. The PTM-Mamba model is the first and only parse tree model (PLM) that can extract distinct and precise latent embeddings from both wild-type and changed protein sequences.
PTM-Mamba Architecture
Amino acids and PTM tokens are both represented by PTM-Mamba, a bidirectional pLM. It expands upon the most advanced structured SSM, Mamba, by incorporating a gated embedding fusion module and a bidirectional module. As an alternative to transformers, recurrent neural networks (RNNs), and convolutional neural networks (CNNs), which model long-range sequential dependency with sub-quadratic complexity, Mamba is a distinct class of structured SSM. As a precursor to the cutting-edge ESM-2-650M model, PTM-Mamba is trained to maintain an understanding of normal amino acids. While PTM tokens are turned into tokens for ESM-2-650M input, wild-type amino acid tokens are fed into ESM-2-650M to collect its output embeddings. PTM-Mamba’s embedding layer receives sequences and automatically handles PTM and wild-type tokens. A novel gating mechanism is proposed to join the ESM-2-650M and PTM-Mamba embeddings, resulting in a final output representation.
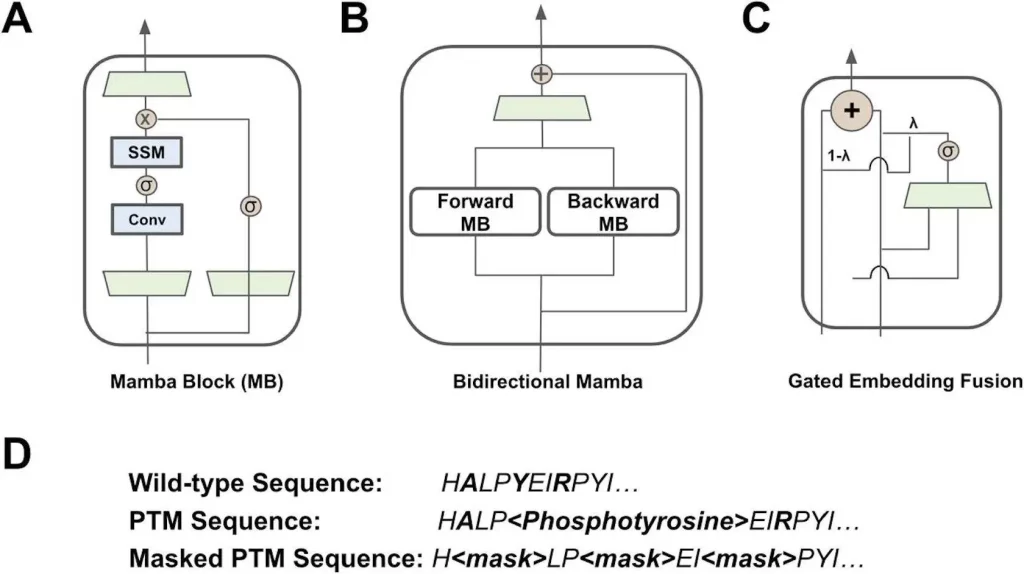
Image Source: https://doi.org/10.1101/2024.02.28.581983
Application of PTM-Mamba
PTM-Aware Embedding Exploration:
PTM-Mamba’s main goal is to accurately and meaningfully depict both unmodified and post-translationally modified protein sequences, recognizing the vital roles that PTMs play in biology and the structural alterations they cause. In a two-dimensional setting, the PTM-Mamba model distinguishes between protein sequences that are wild-type and those that are PTM. The closeness of the embeddings for each wild-type: PTM-modified pair shows that the model can identify the semantic relevance of PTMs while preserving the context integrity of the protein sequence. Additionally supported are the distinct token embeddings produced by PTM-Mamba for PTM tokens as opposed to wild-type amino acid tokens. Spatial proximity characterizes modified residue tokens that pertain to particular PTM classes, like phosphorylation and acetylation. PTM-Mamba shows strong attention to PTM residue tokens, suggesting that the model is primarily concerned with adding PTM residue information to ESM-2 embeddings.
Phosphorylation Site Prediction:
One crucial PTM that is important to apoptotic pathways is PTM-Mamba. The capacity of its embeddings to forecast phosphorylation sites in input protein sequences has been assessed. According to the study, PTM-Mamba performs better than any other model—including ESM-2-650M—in every classification statistic. Its improved performance is true for any length of sequence, indicating that it can be applied to a wide range of protein species. PTM-Mamba has the ability to identify apoptotic pathways because it can be used instead of ESM-2 for PTM-specific property prediction of wild-type sequences.
Non-Histone Acetylation Site Prediction:
The prediction capacity of PTM-Mamba, a PTM with essential functions in DNA repair, protein stability, and gene regulation, was assessed. PTM-Mamba exhibited superior performance over ESM-2-650M across all classification measures, including different input sequence lengths. This demonstrates the value that PTM representations provide to the ESM-2-650M embeddings and encourages their use for other PTM site prediction applications.
Disease Association Prediction of Modified Sequences:
The protein-tissue marker PTM-Mamba is essential for controlling cellular signaling pathways and protein function. On the other hand, these regulatory processes can be altered by erroneous or aberrant PTMs, which can result in illnesses like Alzheimer’s and cancer. Researchers employed the distinct token embeddings of PTM-Mamba to enhance the illness association categorization of changed sequences. Following the creation of an extensive labeled dataset containing PTM sequences, baseline embeddings were used to train a basic MLP classifier. Because ESM-2-650M did better on this task than ESM-2-3B, PTM-Mamba may justify using it. PTM-Mamba exhibited superior performance on pertinent metrics, such as AUROC and AUPRC, indicating the usefulness of its PTM token representations in capturing effects unique to PTMs. This study emphasizes how critical it is to comprehend and mitigate any risks that may arise from improper or abnormal PTM installation.
Conclusion
In order to capture PTM-specific effects in its latent space, this work introduces the first pLM trained to represent both wild-type and post-translationally modified protein sequences. To integrate new PTM residue tokens with ESM-2 embeddings and expand ESM-2’s expressive latent space to accommodate changing protein sequences, a bidirectional gated Mamba block is learned in this manner. Protein sequences having PTM-altered residues encoded at inference time can only be entered into PTM-Mamba. This work demonstrates the strength of the hardware-efficient, powerful Mamba architecture for protein modeling, as well as the potential of SSM primitives for protein language modeling. Sequence-based protein design is made possible by the PTM-Mamba pLM, allowing for the development of binders that identify particular post-translational protein states. By concentrating on the target sequence, this method expands on earlier research in the creation of protein-binding peptides. Subsequently, PTM-Mamba embeddings will be added to the PepMLM architecture to enable binder design to target specific modified proteoforms. The ultimate objective is to provide a level of targeted specificity in drug discovery pipelines by computationally designing and experimentally validating biologics that specifically control changed disease-causing proteins.
Article source: Reference Paper | PTM-Mamba model is available at Hugging Face
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.