While chromosomal synthesis and cloning are expensive and time-consuming processes, CReATiNG (Cloning, Reprogramming, and Assembling Tiled Natural Genomic DNA), a method introduced by researchers at the University of Southern California, presents a viable substitute. With this technique, sections of natural chromosomes are cloned and put together to create synthetic chromosomes. CReATiNG can change a cell’s structure, remove non-adjacent areas, recombine strains, and replace native chromosomes. This new DNA assembly process has the potential to greatly enhance the manufacturing of pharmaceutical drugs, cancer treatments, biofuels, and other biotechnological products.
Understanding CReATiNG
CReATiNG is a technique for creating artificial chromosomes in Saccharomyces cerevisiae using naturally occurring components. Cloning natural chromosomal segments with distinct adapter sequences affixed to their termini—which indicate how these molecules will recombine with each other when they are assembled—is the initial stage in the CReATiNG process. Co-transforming cloned segments into cells and assembling them in vivo through homologous recombination is the second stage of CReATiNG. It is feasible to directly examine the phenotypic consequences of synthetic chromosomes created with CReATiNG by replacing the natural chromosomes in cells.
Significance of Synthetic Chromosome Synthesis
Today, chromosomal synthesis provides answers to basic biological concerns. For instance, answering questions like what is the bare minimum of genes needed by a living cell has long been a mystery. To provide an explanation, scientists created a Mycoplasma mycoides chromosome with just 473 genes that still results in a free-living bacteria that replicates on lab timeframes using design-build-test cycles. In order to create this minimum cell, 428 (48%) of the genes found in M. mycoides naturally have to be removed. Of the genes still present in this minimum cell, 83% were involved in cytosolic metabolism, the cell membrane, or the expression and preservation of genetic information, while the remaining 17% were unknown. The ability to use chromosomal synthesis to comprehend the defining mechanisms behind cellular life and its diversity is demonstrated by the production of this basic cell.
Synthetic chromosomes have only ever been created from scratch, using a combination of in vitro and in vivo methods to gradually assemble tiny synthetic DNA pieces into bigger molecules. De novo synthesis is potent because it permits total reprogramming of the structure and sequencing of a chromosome. De novo chromosomal synthesis, for instance, was utilized to create an Escherichia coli strain that only uses 61 codons by synonymously reprogramming all 18,214 occurrences of three codons. Another example is the strain of the model budding yeast Saccharomyces cerevisiae that the Sc2.0 community creates using de novo chromosome synthesis. In this strain, all transposable elements have been removed, and LoxP sites have been incorporated between genes, allowing the creation of random chromosome rearrangements by Cre recombinase.
The synthesis and assembly of a significant number of DNA fragments required for de novo chromosomal formation restricts its application in biological research. Wider applications of chromosomal synthesis by biologists require lower labor and reagent costs. Creating synthetic chromosomes from cloned natural DNA segments may be a quick and reasonably priced substitute for de novo chromosomal synthesis. With the use of this technique, chromosome synthesis could be used for studies that don’t require whole chromosome reprogramming. Projects that could be made possible, for instance, include tracing the genetic basis of traits that differ between individuals and species, examining the structural requirements of chromosomes, and simplifying chromosomes by methodically removing genetic sequences that are not necessary.
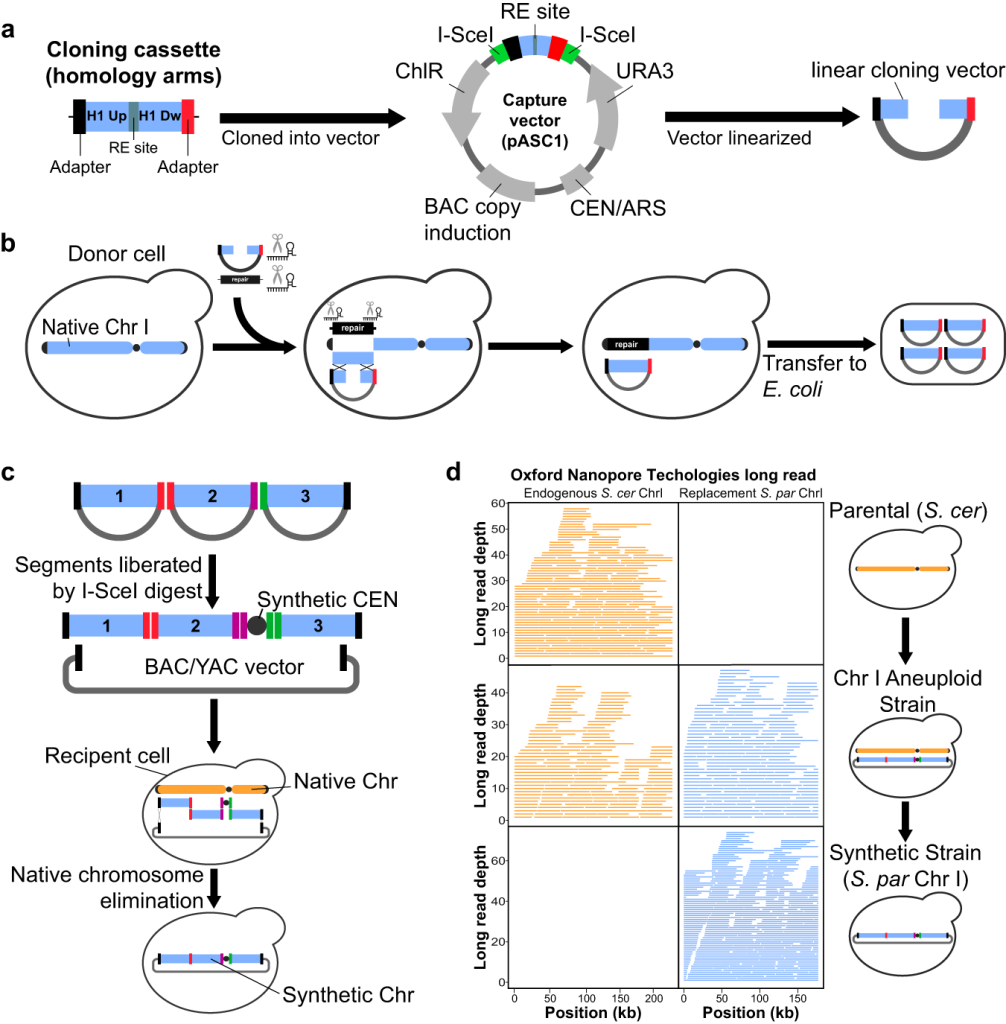
Image Source: https://doi.org/10.1038/s41467-023-44112-2
Application of CReATiNG
- Multiplex gene deletion and chromosome streamlining – Multiplexed deletion could enable the generation of streamlined chromosomes in which many non-essential genetic elements have been eliminated. Such streamlining can facilitate the production of yeast strains with a substantially reduced gene complement, as well as the reorganization of functionally related genes into modules. CReATiNG simplifies multiplexed deletion: segments of a natural chromosome that should be retained can be cloned and assembled, cleanly deleting all intervening parts of a natural chromosome.
- Chromosome restructuring – An issue related to genome function and evolution, chromosomal organization’s structural rules can also be empirically investigated with CReATiNG. Although most of these investigations maintained the order of naturally related genes, recent research indicates that yeast can tolerate a variety of chromosomal configurations. A chromosome’s contents can be reorganized using CReATiNG into particular, non-natural configurations programmed using adapters. Chromosomes having one or more inversions, duplications, deletions, or changes to gene order can be created using CReATiNG.
- Recombination of chromosomes between strains and species – The researchers cloned three ChrI segments from BY and another S. cerevisiae strain, RM11-1a (RM), using CReATiNG to synthesize chromosomes. When the resultant recombinant chromosomes were put together one at a time, the growth rates in both circumstances differed significantly. At both temperatures, the strain with a complete S. paradoxus ChrI grew at the slowest rate; however, at higher temperatures, its divergence from other strains was more pronounced. The three ChrI segments’ individual and combined phenotypic impacts were also assessed in the study, and it was discovered that two of the segments significantly affected growth at 30°C and all three segments significantly affected growth at 35°C. The detection of epistasis between the segments suggests that both additive and epistatic loci have a role in the thermotolerance variations between the species. CReATiNG can offer novel insights into the genetics of phenotypic differences between species, including genotype-by-environment interactions and epistasis, even when the data cannot resolve causative genetic differences.
- A system for cloning and reprogramming natural DNA for assembly – In CReATiNG, natural chromosomal segments are cloned from yeast donor cells, and the cloned segments are then programmable and assembled into synthetic chromosomes in distinct recipient cells. The researchers co-transformed three reagents into donor cells that constitutively express Cas9 to clone a target segment: (1) a linear Bacterial Artificial Chromosome/Yeast Artificial Chromosome (BAC/YAC) cloning vector (pASC1) flanked by homology to the ends of the segment, enabling integration of the segment into the vector in vivo by homologous recombination; (2) in vitro transcribed (IVT) guide RNAs (gRNAs) that direct Cas9 to cut a target segment on each side, excising it from a chromosome; and (3) a repair template that consists of a homology arm-flanked dominant drug marker (KanMX) that enables a cell to reconstruct its broken chromosome by homologous recombination, replacing a segment with a marker.
- Initial assembly of a chromosome using CReATiNG – In order to preserve functional parts, S. paradoxus ChrI, was divided into three segments, each containing about one-third of ChrI, for the CReATiNG cloning and assembly procedure. Due to the use of a synthetic centromere cassette containing CEN6, the native ChrI centromere was not included in the cloning process. Given that they cannot be cloned, subtelomeres and telomeres were left out of the research.
An S. paradoxus CBS5829 strain that expressed Cas9 was utilized for three transformations to facilitate the cloning of the S. paradoxus ChrI segments. After that, the segments, a linear pASC2, and the centromere cassette were co-translated into the BY4742 (BY) reference strain of S. cerevisiae, where they were assembled in their original orientations and order. Conditionally destabilizing and selecting against BY ChrI resulted in the production of the euploid strain (IEY394), which solely contains S. paradoxus ChrI. In order to eliminate a chromosome, an inducible GAL1 promoter was used to disrupt centromere function, and 5-FOA was used to counterselect a URA3 marker on the chromosome. The euploid strain IEY394, apart from a single point mutation in a non-functional region of the centromere cassette, possessed the predicted sequence and structure throughout the genome. To verify that chromosomes synthesized by CReATiNG may be transformed from circular to linear forms, double-strand breaks were introduced near each junction between the chromosome and the cloning vector by linearizing the synthetic S. paradoxus ChrI using Cas9.
Advantages of CReATiNG
- CReATiNG has the advantage of not requiring natural synteny, mating, or meiosis.
- CReATiNG permits the recombination of three or more paternal chromosomes in a single assembly.
- CReATiNG facilitates recombination between a synthetic chromosome and its native counterpart, enabling cells to recover from undiscovered design defects.
Limitations of CReATiNG
- The primary limitation of CReATiNG for chromosomal recombination is its current inability to be implemented throughout the entire genome.
- CReATiNG prevents the need for total chromosome reprogramming, which is necessary for some synthetic chromosome designs.
Conclusion
A novel technique called CReATiNG makes synthetic chromosomes faster and less expensive than de novo chromosomal synthesis by assembling them from natural components. Recombination between a synthetic chromosome and its native counterpart can help organisms recover from unforeseen design errors and facilitate the research of fundamental topics in genetics, genomics, and evolution. In contrast to in vitro techniques such as ExoCET42, CATCH43, and CAPTURE44, CReATiNG’s in vivo cloning strategy exhibits low sequence limitations and is extremely efficient throughout a wide range of target sequences and sizes.
Important biological discoveries can also be made using CReATiNG to construct synthetic chromosomes with intricate patterns, including at least ten segments. Larger numbers of linked, non-adjacent chromosomal areas can be deleted, and it can help reveal natural design principles guiding chromosome architecture and permit fine-scale genetic mapping of heritable traits within and between species. It can also be used in conjunction with de novo chromosomal synthesis to execute projects that might not be possible otherwise.
With the ability to create synthetic chromosomes with a variety of designs and to efficiently relocate genes in the same pathways, complexes, or cellular processes to common genetic modules, CReATiNG democratizes the use of chromosomal synthesis in a wide range of biological research applications.
Article Source: Reference Paper | Reference Article
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.
















[…] CReATiNG Synthetic Chromosomes in Yeast Opens New Possibilities in Synthetic Biology […]