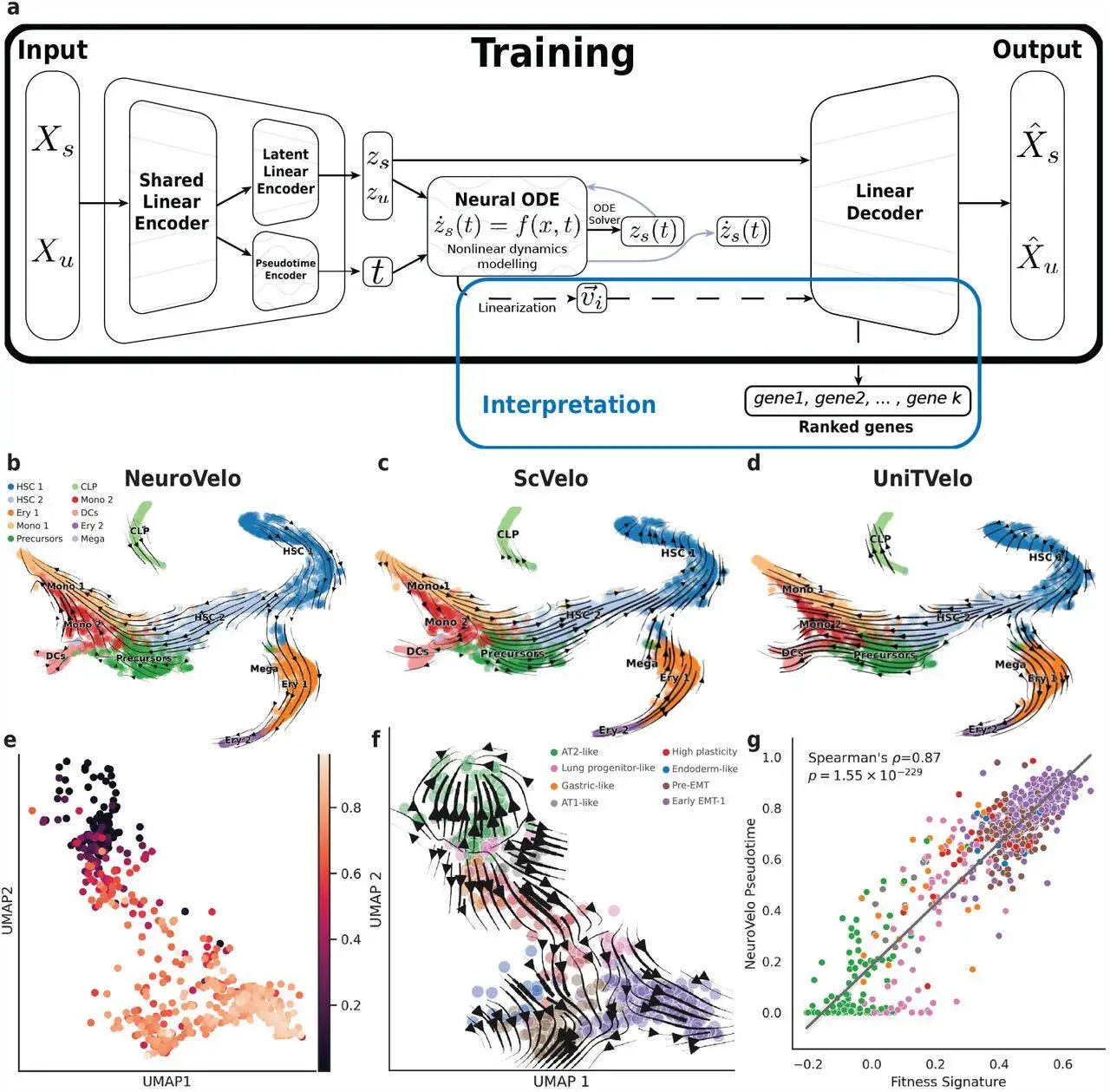
A study conducted at the International School for Advanced Studies (SISSA) in Trieste, Italy, introduced NeuroVelo, a novel method tackling the challenge of reconstructing temporal cellular dynamics from static single-cell transcriptomics. Unlike existing methods, NeuroVelo combines optimal linear projection learning with a non-linear low-dimensional dynamical system, allowing for better interpretation of results and improved predictive power. NeuroVelo excels in identifying genes and biological processes driving temporal cellular dynamics by applying dynamical systems theory. Benchmarked against current methods using single-cell multi-omic data, NeuroVelo proves superior in identifying biological pathways and reconstructing evolutionary dynamics.
Single-cell transcriptome data (scRNA-seq) is revolutionizing our understanding of the diversity and behaviors of cells by acting as a microscope for their internal workings. This amazing technique allows scientists to see inside cells by determining which genes are activated. However, it is somewhat of a challenge to determine from a photograph how cells migrate and alter over time. There are limitations to the existing techniques we employ, such as non-linear dimensionality reduction and RNA velocity. They can make some predictions, but there’s still a lot we don’t know; it’s like trying to read a book with missing pages.
Single-cell transcriptomic (scRNA-seq) refers to the technology used to unravel the heterogeneity and complexity of RNA transcripts within individual cells, as well as to reveal the composition of different cell types and functions within highly organized tissues, organs, or organisms. It has facilitated the temporal transcriptomic recording of single cells, permitting direct mapping of a cell’s journey by progressively analyzing individual cell transcriptomes. But studying changes in gene expression over time of a single cell is difficult with the static snapshots of data because:
- Static single-cell transcriptomic data lacks temporal resolution.
- Biological variability.
- Interpreting the results of cellular dynamics is difficult.
- Limited predictive power.
Although single-cell transcriptomic (scRNA-seq) technologies have revolutionized our understanding of cellular diversity and heterogeneity, their destructive nature limits their ability to capture temporal biological dynamics. RNA-velocity takes advantage of the quite plentiful pre-mRNA readings seen in many scRNA-seq data sets and employs a simple linear ordinary differential equation (ODE) model to determine whether a gene is transcriptionally active or repressed. This concept allows researchers to obtain a glimpse of cellular dynamics even in static data sets. Despite the fact that applications and extensions of the RNA velocity framework have flourished in recent years, both the biochemical foundations and the biological interpretation of RNA velocity have been called into question. The researchers introduced a new method called NeuroVelo to overcome the above drawbacks. NeuroVelo is a method that couples learning of an optimal linear projection with a non-linear low-dimensional dynamical system. Using dynamical systems theory, NeuroVelo can identify genes and biological processes driving temporal cellular dynamics.
Understanding NeuroVelo
NeuroVelo is a novel approach to inferring cellular dynamics from scRNA-seq data that is more easily interpretable and highly effective. By combining ideas from Neural Ordinary Differential Equations (ODE) and RNA velocity, NeuroVelo is formed. NeuroVelo aims to identify genes and biological processes that drive temporal cellular dynamics. This method is more interpretable and effective for deducing cellular dynamics from single-cell transcriptomic data.
Two recent scRNA-seq data sets are used in this study to test NeuroVelo: a human bone marrow hematopoiesis data set and a CRISPR based lineage barcoded mouse cancer data set. In the bone marrow data set, NeuroVelo was able to capture the correct dynamics of early differentiation. In the results of the mouse cancer data set, NeuroVelo produces a wide reconstruction of both pseudo-time and velocity fields. NeuroVelo achieves the strongest Spearman correlation between estimated pseudo-time and fitness signature compared to other methods like RNA-velocity and PhyloVelo.
To put NeuroVelo to the test, the researchers used a single-cell Multiome dataset (nuclear transcriptome + chromatin accessibility – ATAC) of patient-derived colorectal cancer organoids treated with various medicines. A colorectal cancer clinical study was used to create the organoid line. The ancestral organoid line was subjected to three distinct treatment regimens: an AKT inhibitor (capivaserib), a MEK inhibitor (trametinib), and lastly, a combination of the two. As the study focuses on interpretability, the researchers restricted comparisons to lists of high-velocity genes provided by scVelo and UniTVelo. Other deep-learning-based methods are more difficult to understand since they combine non-linear projections/embeddings with non-linear dynamics. NeuroVelo, on the other hand, is based solely on a low-dimensional non-linear dynamical system that can be studied using traditional spectrum methods. The resulting eigenvectors can then be linearly embedded in the original gene space, yielding a list of genes that can be interpreted using traditional methods. NeuroVelo obtains a more distinct evolutionary trajectory compared to scVelo and UniTVelo approaches.
In another case, researchers investigate the FoxO pathway, which has been found by both neuroVelo and scVelo. NeuroVelo identifies a broad gene collection with a high enrichment score, whereas scVelo identifies high-velocity genes in the FoxO pathway that do not exhibit especially coordinated behavior, yielding an overall low enrichment value.
The researchers proceeded to focus on the ten most essential genes discovered using various methodologies. It should be emphasized that the three approaches identify nearly fully non-overlapping sets of genes (just one gene is shared by NeuroVelo and UniTVelo, and only one gene is shared by NeuroVelo and scVelo). The results from the experiment show that NeuroVelo expression levels are higher than those found with scVelo or UniTVelo. Specifically, scVelo finds genes with very low expression levels and thus unknown biological importance. Researchers linked the expression of the genes discovered by NeuroVelo with the chromatin accessibility of their respective genomic areas to seek more biological validation of these gene sets. This was significantly greater than the other methods for the BNIP3 gene, indicating a strong link between the two measurements. These findings show that NeuroVelo can extract biological dynamics from complex single-cell transcriptomic data in an accurate and interpretable way in terms of the underlying genes driving the dynamics. NeuroVelo, according to the researchers, will thus become a key tool in characterizing biological processes in single-cell investigations.
Advantages of NeuroVelo
Using single-cell multi-omic data, the NeuroVelo is compared to many existing approaches. According to the findings, NeuroVelo can beat rival technologies in identifying biological pathways and reconstructing evolutionary dynamics. This demonstrates NeuroVelo’s exceptional accuracy and interpretability in collecting complicated biological dynamics from single-cell transcriptome data.
Conclusion
In the future, NeuroVelo could be a game-changing approach for inferring temporal cellular dynamics from static single-cell transcriptome data. The capacity of NeuroVelo to deliver precise and interpretable insights into cellular dynamics has incredible potential to significantly improve our understanding of cellular behavior and aid in the development of novel therapeutic approaches.
Article Source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.