The effective paradigm of molecular linkers in drug discovery is crucial for obtaining relevant candidate molecules in early-stage drug development. In this study, researchers present DiffLinker, a three-dimensional conditional diffusion model for molecular linker design that is E(3)-equivariant. A synthetically available model called DiffLinker generates molecules from disconnected fragments that include all of the original fragments. It can automatically count the number of atoms in the linker and its attachment locations to the input fragments. It can link an infinite number of fragments. On benchmark datasets, the model performs better than other approaches, producing more varied and readable molecules. It can effectively generate legitimate linkers conditioned on target protein pockets, according to experimental testing conducted in practical applications.
Introduction
Drug design is difficult because there are thought to be more than 1060 pharmacologically significant structures. Fragment-based drug design (FBDD), which begins with smaller molecules containing no more than 20 heavy atoms, is an effective strategy. Computationally identifying fragments that interact with a protein pocket is a more cost-effective and efficient screening method than experimental approaches. It is then necessary to merge the pertinent fragments into a single, connected chemical complex after they have been found and docked to the target protein. The effective design of relevant and potent compounds depends on the geometries of recognized fragments, as shown by applications such as scaffold hopping, PROTAC design, and FBDD. The affinity of the produced chemical leads can be greatly enhanced by the configuration of the protein pocket during the linker design procedure. The challenge of connecting fragments arranged in a three-dimensional (3D) context is tackled in this work, constraining the design procedure to the target protein pocket.
Molecular linker design was first approached computationally through database searches and computationally demanding physical simulations. The use of machine learning techniques to produce various linkers more quickly is becoming more and more popular. Current methods include autoregressive models and syntactic pattern recognition, which only work with SMILES and take into account the three-dimensional positions and orientations of input fragments. These techniques can only join pairs of fragments and are not equivariant with regard to atom permutation. There isn’t a computational approach for designing molecular linkers that takes the target protein pocket into account yet.
Understanding DiffLinker
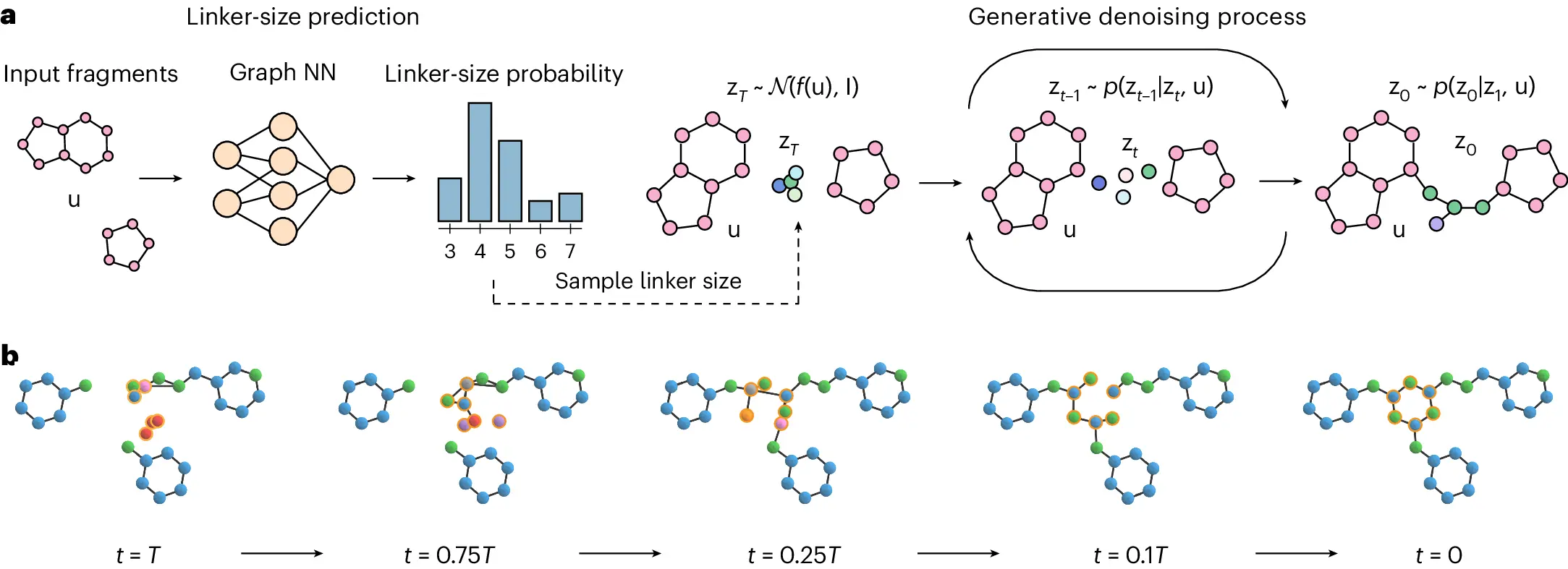
DiffLinker is a 3D atomic point cloud conditional diffusion model that creates molecular linkers for input fragments. Using a neural network conditioned on the input fragments, the model produces the size of the prospective linker, samples the initial linker atom types and positions, and updates them repeatedly. A single linked molecule is formed from the input fragment atoms and denoised linker atoms.
The desirable qualities of DiffLinker are as follows:
- It produces linkers with no preset size, does not require information on the attachment atoms,
- It is equivariant to translations, rotations, reflections, and permutations,
- The number of input fragments does not restrict it.
A cutting-edge generative technique called DiffLinker shows exceptional effectiveness in creating chemically meaningful linkers between pairs of fragments. Drug design pipelines can benefit from this strategy since it produces synthetic accessibility and drug-likeness. When it comes to the chemical variety of the created linkers, DiffLinker performs better than other approaches. It can effectively connect more than two fragments, something that other techniques are unable to do. The process can be tailored to the target protein pocket while taking into account the geometric limitations given by the nearby protein atoms. Three case studies illustrate its integration into fragment-based ligand design targeting heat shock protein 90 (Hsp90) and inosine 5′-monophosphate dehydrogenase (IMPDH) as well as scaffold hopping for enhancing selectivity for c-Jun N-terminal kinases (JNKs) are presented to illustrate its relevance in real-world drug design applications. The first approach that takes pocket information into consideration and is not constrained by the quantity of input pieces is DiffLinker. The ultimate objective is to give professionals a useful tool for creating molecular linkers in practical drug design situations.
DiffLinker’s Approach
The researchers test the approach on four benchmarks under various conditions:
- The paper presents DiffLinker’s performance on the ZINC and CASF datasets, which only have pairs of fragments that need to be joined. Scientists then present a new dataset that is based on GEOM molecules and has two or more distinct fragments in each entry. Researchers test many iterations of the technique for each of the three sets: known or unknown anchor sites and predefined or sampled linker sizes.
- Researchers evaluate DiffLinker’s capacity to create pertinent linkers when the protein pocket is present. Researchers provide an additional dataset based on Binding MOAD for that purpose. In addition to the common metrics employed in the earlier benchmarks, the study counts the number of steric collisions that occur between the produced linkers and the neighboring protein atoms.
- The study presents the usefulness of DiffLinker in scaffold hopping to increase JNK selectivity as well as in the fragment-based design of Hsp90 and IMPDH inhibitors.
Conclusion
A new 3D conditional diffusion model for molecular linker design called DiffLinker has useful properties that can help speed up the creation of drug candidates by utilizing FBDD techniques. Nevertheless, the method’s poorer validity stems from the raw point clouds it generates, which OpenBabel processes in order to compute covalent bonds. Unlike other approaches, which build bonds and use valency rules at every generational step, the approach is distinct. Although DiffLinker is able to efficiently learn basic chemistry from unprocessed geometric data, it can still be improved by adding edge features, producing chemical bonds with atom kinds and coordinates, and adding information on covalent bonds. Compared to alternative linker design techniques, DiffLinker generates more synthetically accessible compounds, as evidenced by the current work, highlighting the significant attribute of high SA in sampled molecules. Though the distribution of linkers in PROTACs varies greatly, there is still an opportunity for development in terms of producing suitable linkers for PROTAC-like molecules. Retraining the model with more appropriate PROTAC data is recommended to enhance DiffLinker’s performance in PROTAC design. Although DiffLinker is primarily concerned with designing molecular linkers, it can also help with other phases of fragment-based drug discovery.
Article Source: Reference Paper | DiffLinker’s source code is freely available on GitHub
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.