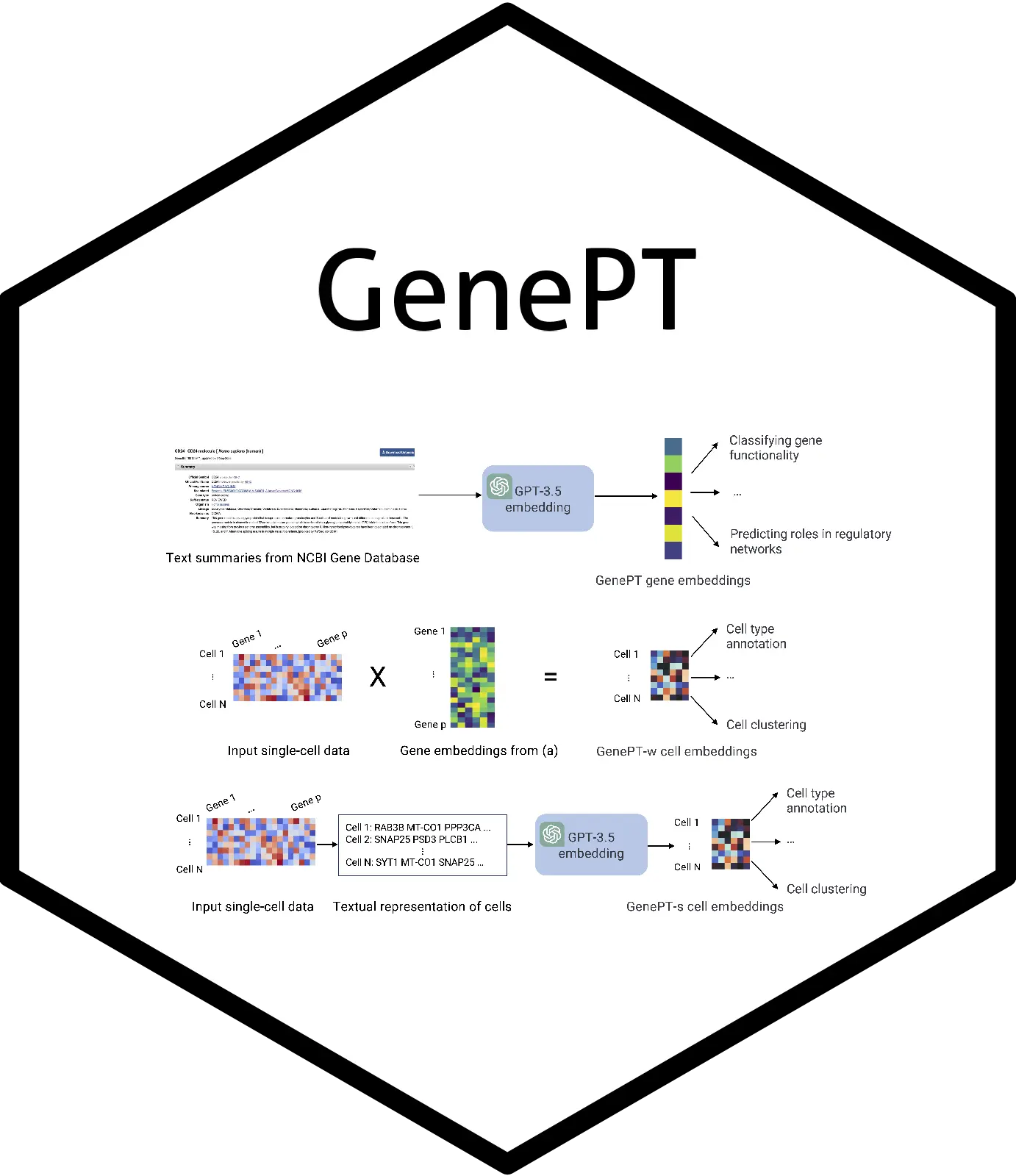
Large-scale gene expression data is used by Geneformer and scGPT models to build fundamental single-cell biology models. These models implicitly learn gene and cellular activities from millions of cell expression profiles, necessitating labor-intensive training and considerable data curation. Here, researchers from Stanford University use gene embeddings from the literature leveraging ChatGPT to investigate a much easier solution. In order to create gene embeddings, GenePT uses the GPT-3.5 and the NCBI text descriptions of specific genes. GenePT is an effective technique for creating single-cell embeddings by averaging gene embeddings according to the expression level of each gene. It performs on par with or better than Geneformer and other models, and it is user-friendly and efficient. GenePT generates sentence embeddings for every cell by sorting gene names according to expression level. It shows that a straightforward and efficient route for biological foundation models is the large language model embedding of literature.
Introduction
The foundation models Geneformer and scGPT are made to learn gene and cell embeddings for further investigations. Using a deep learning architecture, these models link input genes and cells to a high-dimensional embedding vector and collect large datasets of single-cell gene expression for pretraining. A small amount of task-specific data can be used to refine the model and improve its predictive power for downstream tasks. However, this method does not make use of literature or prior knowledge about a gene; instead, it merely extracts embeddings from gene expression datasets. This has drawbacks, including the time and computer resources needed for the extensive gathering and processing of single-cell transcriptomics data. Furthermore, there is a high dependence on the signals from derived embeddings on gene expression data, which may result in sample inefficiency and unsatisfactory outcomes in some applications.
This work investigates the viability of encoding the biology of genes and cells using natural language as a substitute, supplemental method. GPT-3.5 and GPT-4 are two examples of large-language models (LLMs) that have shown exceptional comprehension, reasoning, and production skills in biomedical material. It is proposed that more accurately capturing the underlying biology may be achieved by LLM-derived embeddings of gene summaries and functionality, which are frequently selected from a wide range of experiments and studies.
Understanding GenePT
GenePT is a method that depicts genes and cells in biologically driven tasks by utilizing OpenAI’s ChatGPT text embedding API services. In a variety of jobs, it performs better than and occasionally better than specially built models like Geneformer. Compared to single-cell RNA-seq-based foundation models, GenePT performs better on biological tasks, requires less money for pretraining and curation, and is easier to use when creating gene and cell embeddings. It combines expression-based representations in an intriguing new way by utilizing LLM-based embeddings.
Applications of GenePT
- Capture Gene functionality – GenePT embeddings have been assessed and contrasted in terms of how well they can encode gene activities. The study discovered that because the functionality is commonly present in NCBI gene summaries, GenePT embeddings preserve important biological information. With an accuracy of 96% overall, the predicted functional class matches the genuine annotation quite well. Even with a more complex deep neural network, GenePT embeddings outperform other embeddings obtained from single-cell foundation models in terms of performance. Additionally, the predictive power of GenePT gene embeddings for protein-protein interactions (PPI) was assessed. A possible future step is to integrate GenePT embeddings with protein embeddings learned from 3D structures or protein language models. The results indicate that GenePT’s literature-based embedding contains information relevant to gene and protein interactions.
It has been discovered that gene programs, including those created by GenePT, exhibit functional diversity and variable expression patterns in many cell types. This is consistent with known biological information, such as the discovery of gene sets that make up the CDC and IFI families. Gene programs are obtained from datasets of human immunological tissue and are subjected to a “zero-shot” analysis, which reveals that different cell types selectively activate these programs. This is consistent with known biology and shows how well GenePT-inferred gene programs capture functional categories that are relevant to biology.
- Do Accurate predictions in chromatin dynamics and dosage sensitivity – Using datasets from their literature, Theodoris et al. studied network dynamics between dosage-sensitive and dosage-insensitive TFs, bivalent and non-methylated genes, Lys4-only-methylated and non-methylated genes, and long- and short-range TFs. In order to predict the functions of genes in network dynamics, they employed Geneformer, an improved transformer model. Despite Geneformer’s larger pretraining dataset and more complex classification head, GenePT embeddings regularly produced competitive results, occasionally outperforming it. Similar outcomes to random guessing were also seen with random embeddings. The findings imply that overfitting of the model or a large embedding dimension are not the only reasons for the GenePT’s performance.
- GenePT cell embeddings capture biological variations – The study assesses how well single-cell datasets can capture biology using cell embedding techniques like GenePT. It measures the degree of agreement between biological annotations and k-means clustering labels derived from GenePT-w, GenePT-s, pre-trained Geneformer, and scGPT embeddings. In terms of AMI and ARI metrics, GenePT-s performs better than both GenePT-w and Geneformer embeddings and the concordance measured by these metrics is determined to be in accordance with GenePT-s. Biological changes corresponding to two major single-cell foundation models are captured by GenePT cell embeddings. Concordance with cell types and annotations, however, is a limited indicator of embedding’s usefulness. Additionally, the study discovered that two of the most effective techniques in this situation are scGPT and GenePT-w, which routinely exceed Geneformer in terms of prediction accuracy.
Future Work
Future research regarding GenePT should be focused on expanding its dynamic and context-dependent characteristics, integrating information unique to tissues, diseases, and marker genes. Important directions for future research include utilizing dimension reduction techniques for compact representations and integrating various embeddings across single-cell foundation models. It would also be possible to investigate GenePT’s performance in drug-gene interactions and perturbation predictions. Furthermore, protein sequence modeling and genome-wide association studies, two other biological disciplines, might benefit from using natural language descriptions with LLMs embedding.
Conclusion
The NCBI is a useful tool for learning about genetic and cellular functions, thanks to the development of GenePT, a straightforward but efficient method that represents genes and cells using GPT-3.5. This method works well for identifying gene functionality groups and predicting interactions between genes, among other applications. However, because the existing GenePT system relies solely on publicly available gene summaries and descriptions, it might miss some lesser-known features that aren’t listed in databases like NCBI. Furthermore, certain tissues and cell types may not benefit from GenePT embeddings, which presents difficulties in capturing the dynamic and context-dependent roles that genes and cells play. The language models used namely GPT-3.5, limit the efficacy of the embeddings. Optimizing the language models may help improve comprehension of the domain-specific terminology used in genomics.
Article source: Reference Paper | GenePT is available on GitHub
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.