Users without prior Machine Learning (ML) experience have difficulties when it comes to training and implementing deep learning models. Regardless of prior experience with machine learning, SaprotHub, introduced by the researchers of Westlake University, provides a simple platform for training, using, and exchanging protein models, encouraging cooperation among biologists. All it takes is a few clicks to get started. Saprot, also known as ColabSaprot, is essentially a near-universal protein language model that performs exceptionally well on a variety of protein prediction tasks.
Introduction
As the fundamental units of life, proteins are essential for a wide range of biological functions and are key players in advances in genetics, medicine, and pharmacology. Despite their importance, the scientific community still finds it extremely difficult to solve the complex structure and function of proteins. With its success in CASP14, AlphaFold2 has opened a new chapter in structural biology by successfully predicting protein structures at the atom level. Large-scale protein language models are driving major advancements in the prediction of protein function.
ProtTrans, UniRep, ProGen, and ProtGPT2, are examples of ESM models that have demonstrated effectiveness in their particular duties. However, developing and implementing these machine-learning models for proteins presents difficulties for researchers. These difficulties include choosing appropriate model architectures, handling coding quirks, preparing datasets, training model parameters, and assessing results. Non-ML researchers are frequently discouraged from actively engaging in this field because of its complexity. ColabFold is an innovative project that uses Google Colaboratory to host AlphaFold2 predictions in order to make protein folding accessible to researchers from all backgrounds. Nevertheless, ColabFold does not include protein function prediction; instead, it is restricted to protein structural tasks and concentrates on model inference.
What is SaprotHub?
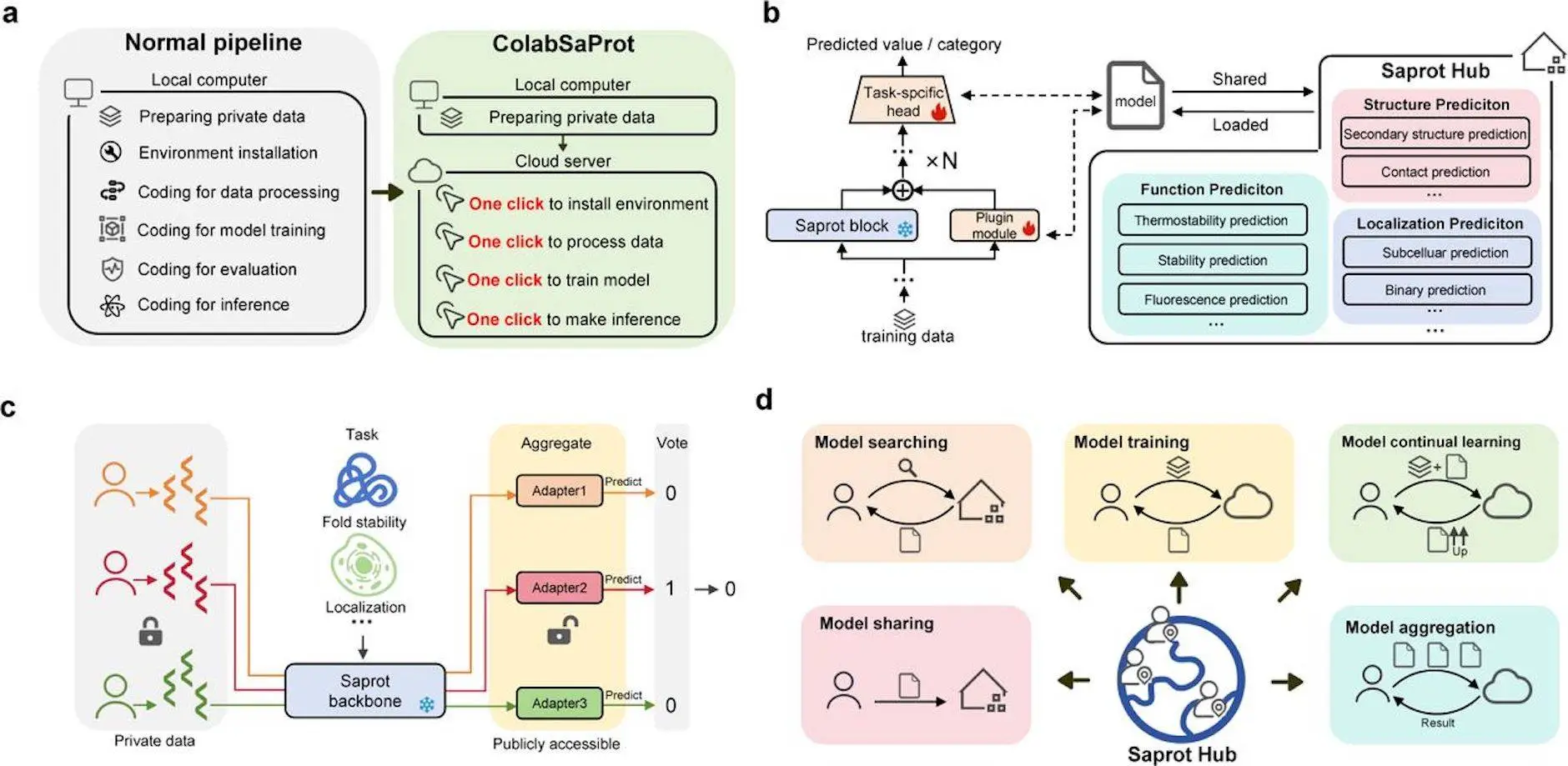
SaprotHub is a user-friendly tool used by researchers to predict protein function. With the help of this platform, researchers without extensive ML or coding experience can build and train their own models. Rather than being limited to particular functions, SaprotHub supports a wide variety of protein function predictions. By pooling resources and efforts, the “Open Protein Modelling Consortium (OPMC)” hopes to facilitate the sharing of independently trained models among researchers in the protein community. This goal is intended to be realized through SaprotHub, which represents a major step towards the realization of OPMC.
SaprotHub is composed of three components:
- Saprot, a universal protein language model;
- ColabSaprot, which uses adapter learning techniques on the Colab platform to fine-tune and infer Saprot;
- An open community store allows for collaboration, sharing, and retrieval of refined versions of Saprot models.
Advantages of SaprotHub
SaprotHub successfully solves a number of significant obstacles by combining adapter-based fine-tuning approaches, Colab cloud computing technologies, and inventive integration of foundation protein models. These include the vast parameter scale of the model, which makes it difficult to share and aggregate, the possibility of catastrophic model forgetting from ongoing learning, and the requirement to safeguard biologists’ proprietary data.
SaprotHub Methodology
The Structure-Aware (SA) protein representation technique, which is based on amino acid sequences or 3D coordinate structural representations, is introduced by the protein foundation model SaprotHub. For proteins, it makes use of the SA vocabulary (SAV), which covers all possible pairings of AA and 3Di types. Similar to ESM-2, Saprot’s neural network architecture uses a bidirectional Transformer block. In the pre-training phase, Saprot predicts the full form of a few randomly chosen SA tokens that have been partially hidden. Like ESM-2 and AlphaFold3, SaprotHub was trained on about 40 million protein SA sequences over a three-month period utilizing 64 NVIDIA 80G A100 GPUs. AlphaFold2 is used to anticipate the protein structure that will be utilized to represent the SA sequence during training.
Comparing Saprot to ProtBert and ESM-2, a protein language model, shows superiority in more than ten important tasks. Saprot is still competitive even without the ability to input protein structures. In three challenges that anticipate zero-shot mutational effects, it performs better than ESM-2 beyond more than 60 well-known baselines, such as ESM-2, EVE, ESM-1v, MSA Transformer, ESM-IF, DeepSequence, and UniRep. Saprot also holds the top spot on the publicly accessible blind ProteinGym leaderboard.
About Saprot
A flexible protein language model, Saprot may be used for a variety of applications such as protein inverse folding, zero-shot prediction, and supervised prediction. It can manage a broad range of regression and classification tasks, including interactions and similarities between proteins. Without further fine-tuning, Saprot can predict viral mutation fitness and enzyme activity in zero-shot prediction scenarios. It is also very good at finding driver mutations and forecasting the impact of disease variants. Saprot isn’t meant to be generative in the traditional sense, but it can still be quite good at folding proteins inversely, which opens up possibilities for community cooperation and co-construction in this context.
Understanding ColabSaprot
Researchers may perform training sessions with ease thanks to ColabSaprot, a platform that incorporates pre-trained Saprot as its core model. With no hardware limitations, its strong architecture guarantees resource availability and scalability, enabling effective model training. Researchers can alter the code, data, and training hyper-parameters to meet their own requirements. All functions included in the Saprot framework can be directly utilized with ColabSaprot, including protein inverse folding and zero-shot mutational effect prediction. Compared to fine-tuning all Saprot parameters, a parameter-efficient fine-tuning method is presented that enables almost the same accuracy. This architecture increases the effectiveness of optimization and encourages teamwork in the creation of different Saprot models among researchers.
Conclusion
SaprotHub is a platform that enables biologists to share and co-build trained Saprot models within the biology community. It offers advanced functionalities like model saving, searching, sharing, continual learning, and aggregation, allowing biologists to train models based on shared models from peers. This is particularly beneficial when data availability is limited, as fine-tuning on a superior pre-trained model improves predictive accuracy. As more biologists join SaprotHub, multiple Saprot models will be submitted for the same protein function, enhancing predictive performance. The open-sourced code for both Saprot and ColabSaprot enables other protein foundation models to adopt similar approaches or become part of SaprotHub.
Article Source: Reference Paper | Saprot is available under an MIT license on GitHub. Fine-tuned models are available at HuggingFace.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.