When compared to other mental conditions, posttraumatic stress disorder (PTSD) has a lower discoverability rate. A meta-analysis encompassing many ancestries, including European, African, and Native American, revealed 95 relevant sites across the genome and 43 putative causative genes. These genes are categorized as developmental factors, immunological or endocrine regulators, neurotransmitter and ion channel synaptic modulators, and synaptic structure and function genes. These findings expand on our knowledge of the neural pathways pertinent to the pathophysiology of PTSD and provide new avenues for research, all of which contribute to a better understanding of the disorder.
Introduction
After exposure to a traumatic event, some people may experience chronic symptoms of posttraumatic stress disorder (PTSD), which include intrusive thoughts, hyperarousal, avoidance, and unfavorable changes in mood and cognition. The prevalence of PTSD among individuals who have experienced trauma is 5.6% globally; rates are higher in those who have experienced severe levels of trauma and specific forms of trauma, such as survivors of assault and combat. For many, PTSD is a chronic illness that has a significant negative impact on both the individual’s quality of life and the economy of the community.
Preclinical research, many of which concentrate on the brain’s fear systems and some of which are being applied to patient PTSD investigations, is making significant strides towards understanding the biology of PTSD. Studies on human neuroimaging suggest that the brain’s fear circuitry may be dysfunctional. This may include deficiencies in the anterior cingulate cortex and ventromedial prefrontal cortex’s ability to top-down modulate the amygdala.
The development and maintenance of PTSD have been linked to disruptions in the HPA axis and glucocorticoid-induced gene expression, according to neuroendocrine research. New targets are required for the prevention and therapy of PTSD, as there are still many unanswered concerns regarding its pathogenesis.
Drawbacks of Previous Studies
Although genetic and twin studies have shown that PTSD risk is partially influenced by genetic factors, the precise genetic architecture of PTSD is only now beginning to be understood as a result of the availability of very large meta-analyses of genome-wide association studies (GWAS). The Psychiatric Genomic Consortium for PTSD (PGC-PTSD) and the VA Million Veterans Programme (MVP) have conducted recent research that has enhanced understanding of the genetic complexity of PTSD as a highly polygenic condition. This research found up to 15 PTSD risk loci, which were inconsistent among datasets and required even higher sample sizes despite sample sizes of over 200,000 individuals. Furthermore, the X chromosome, which makes up 5% of the human genome and may be especially significant given sex variations in the prevalence of PTSD, was not examined in this research.
Furthermore, the ability of GWAS to find reliable treatment candidates has been somewhat limited until now. Additionally, it is well known that PTSD is frequently genetically and comorbid with other mental health disorders (like major depressive disorder [MDD]; attention deficit hyperactivity disorder) as well as physical health conditions (such as obesity and cardiovascular disease). However, the ability of studies to identify shared and disorder-specific loci and connect them to underlying biological systems has been limited to date.
Significantly, the ability of earlier GWAS to extrapolate their results to non-European ancestries is severely constrained. Polygenic risk scores (PRS) have been the subject of a recent study on PTSD, which suggests that these measures may be useful in research, but their cross-population transferability is frustratingly limited. Recent developments in the genetics of PTSD run the danger of exacerbating treatment and research gaps unless they are expanded to include more ancestries. Given the disproportionately high rates of trauma and PTSD experienced by groups of African, Native American, and Latin American descent, this disparity is especially concerning in the United States.
Key Features of the Study
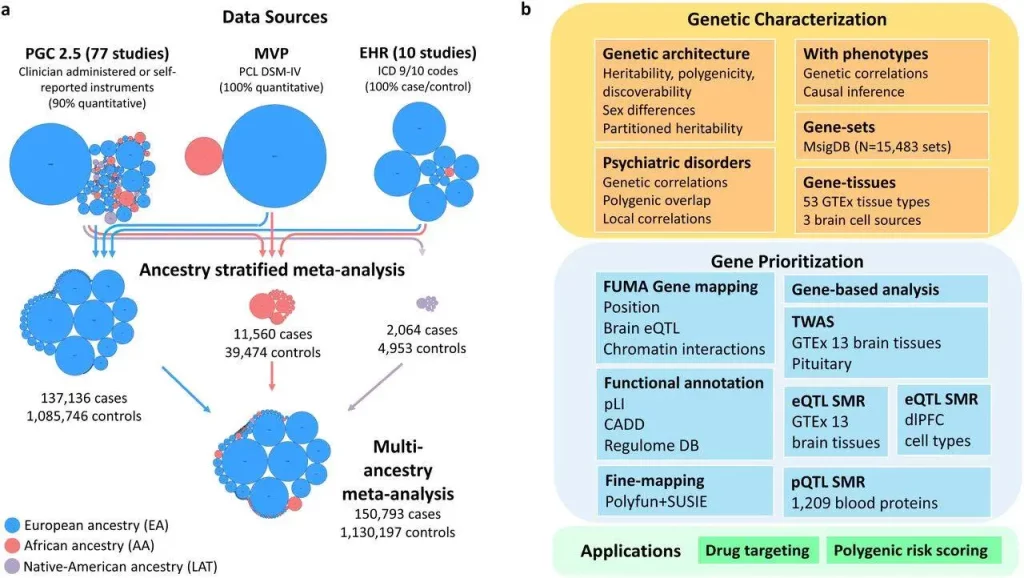
- In this analysis, researchers conduct a multi-ancestry meta-analysis of GWAS data from samples with European ancestry (N = 137,136 cases and 1,085,746 controls), African ancestry (N = 11,560 cases and 39,474 controls), and Native American ancestry (LAT) (N = 2,064 cases and 4,953 controls), including X chromosome analyses.
- Researchers do this by synthesizing data from 88 studies. Following up on the results of the GWAS, they investigate both global and local heritability, use transcriptome data to infer the involvement of brain areas and neuronal systems, characterize genetic effects that are shared with comorbid diseases, and prioritize a set of 43 putatively causal genes using multi-omic data.
- Finally, researchers make use of this data to pinpoint possible research avenues for PTSD therapy studies in the future.
Findings of the Study
- Tissue and cell-type enrichments demonstrated the role of interneurons and the cerebellum in PTSD risk, in addition to other brain regions that are typically linked to PTSD.
- Large-scale postmortem transcriptome investigations of PTSD consistently show differential expression of interneuron markers in prefrontal cortical tissue and amygdala nuclei. These studies also link PTSD to structural changes in the cerebellum.
- Using brain transcriptome data, researchers used TWAS and SMR to investigate the causative genes present in the enriched tissues and cell types. The detected signals were focused in specific GWAS locations, such as 17q21.31, whose inversion area has been linked to alterations in brain structure and function as well as a variety of mental disorders. In both neuronal and non-neuronal cell types, the top causative genes were KANSL1, ARL17B, LINC02210-CRHR1 (encoding a fusion protein with CRHR1), and LRRC37A2. KANSL1 is essential for the development of the brain.
- Moreover, the first single-cell transcriptome research of PTSD demonstrated the involvement of immunological and glucocorticoid response in neurons and validated neuronal, excitatory, and inhibitory changes in 17q21.31, with top modifications in ARL17B, LINC02210-CRHR1, and LRRC37A2.
Conclusion
In this study, researchers examined data from more than a million participants in the biggest PTSD GWAS to date, and the researchers found 95 independent risk loci overall across analyses—a five-fold increase over the most recent PTSD GWAS. In contrast to earlier GWAS for PTSD, they found 80 new PTSD loci and validated 14 of the 24 loci. In psychiatric GWAS, variant finding exhibits a sigmoid curve that quickly increases as the sample size above a certain threshold. This analysis of PTSD surpasses that turning point, signifying a substantial advancement in the genetics of PTSD. The findings also offer fresh, functional insights and a more thorough characterization of the genetic architecture of PTSD by utilizing complementing research approaches. In an EA meta-analysis, researchers found 81 loci linked to PTSD; when they expanded to trans-ancestry studies, they found 85 loci. Even though these findings indicate intriguing potential target genes and mark a significant advancement in the field of PTSD genetics, more funding for the collection of data from underrepresented populations with a variety of ancestries is required in order to identify additional risk variables and produce more robust and equitable PRS.
Article Source: Reference Paper | Reference Article | Analysis code is available publically on GitHub and Zenodo.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.
















