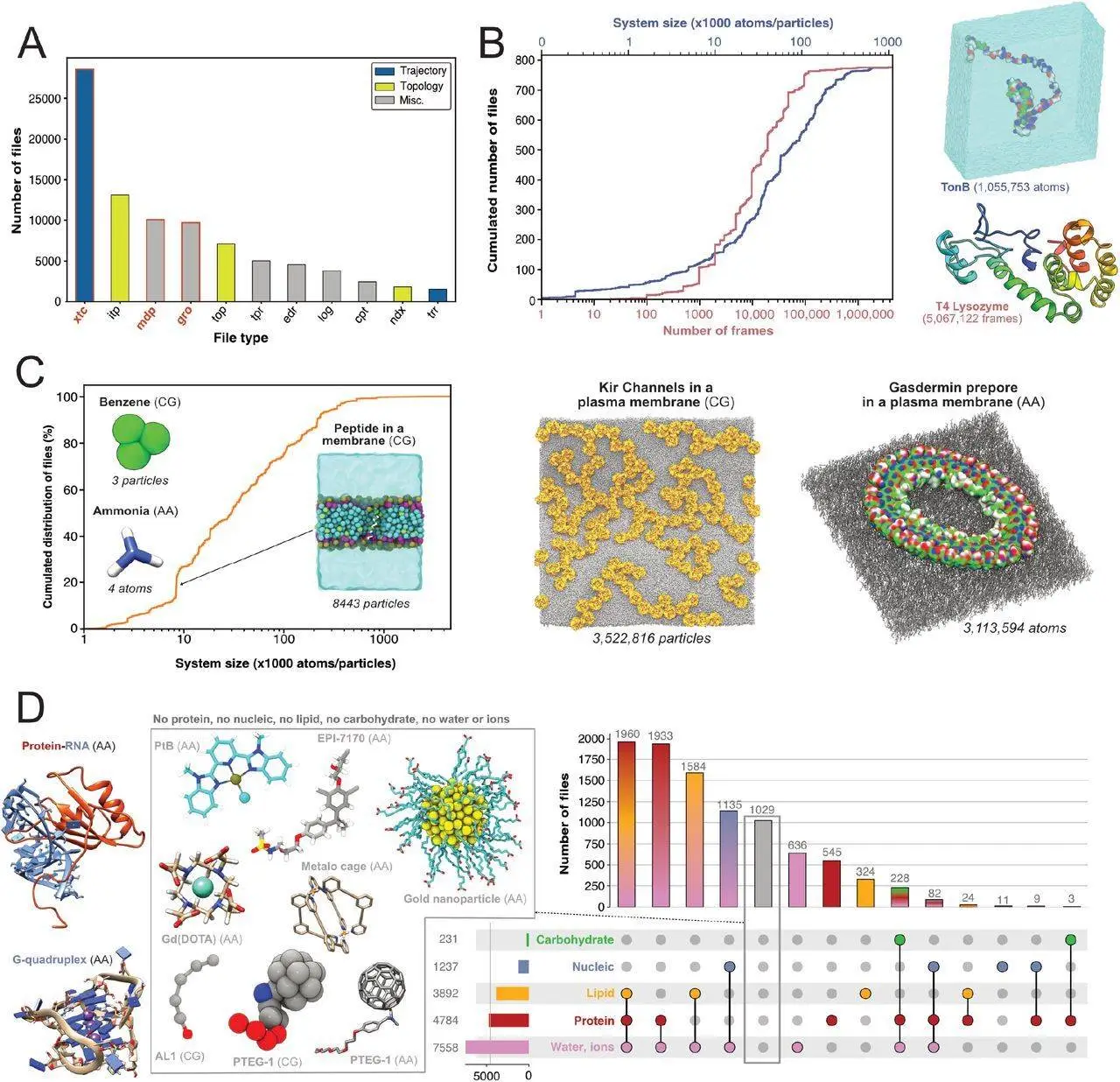
Molecular dynamics (MD) files have accumulated in generalist data repositories due to the rise of open science and the lack of a worldwide repository specifically for MD simulations. This data is known as the “dark matter” of MD, which is technically accessible but not indexed, curated, or readily searchable. Researchers discovered and indexed over 250,000 files and 2,000 datasets from Zenodo, Figshare, and Open Science Framework by using an innovative search method. Researchers demonstrate the possibilities provided by mining publicly accessible MD data, concentrating on files generated by the Gromacs MD program. Researchers could identify systems with particular chemical compositions, characterize crucial MD simulation parameters like temperature and simulation duration, and distinguish between all-atom and coarse-grain model resolution. Using the information, researchers deduced from this study and proposed a prototype search engine, MDverse, to examine the MD data.
Introduction
Research data volume has expanded dramatically in biology due to the development of high-throughput experimental tools and effective computational methods. The proliferation of research data has been fueled by the Open Access movement, which has made research findings publicly available and free of charge. In an effort to reuse research data and promote scientific reproducibility, the FAIR principles were developed to organize the sharing of this data. As a result, new artificial intelligence (AI) applications have emerged. One example is AlphaFold, which allows one to create a structural model of any protein based on its sequence. However, the availability of clean, well-annotated public databases with protein sequences and structures was the only thing that made the construction of AlphaFold feasible. A community-driven compilation of NMR data in the Biological Magnetic Resonance Data Bank allowed for accurate predictions of NMR chemical shifts and chemical-shift-driven structure determination. If the data is well chosen and widely accessible, one might envision new possibilities for AI and deep learning to reuse earlier study findings in various domains.
Understanding MD and its data
In the study area known as molecular dynamics (MD), dynamic processes—from material science to biological phenomena—are understood via the use of simulations. Various experimental methodologies are complemented by these simulations, which reveal motions at details and timescales imperceptible to human sight. When massive molecular systems are modeled or biassed sampling techniques are used, a significant volume of MD data is produced. These models are usually utilized for a single publication, but storage is necessary for researchers to examine the simulations again. Comparing storage costs to the resources needed to produce simulation data reveals that it might be a treasure trove of data for investigators. The data produced by MD simulations must be made both theoretically and practically accessible to the scientific community in the age of open and data-driven science. The endeavor of the MD community to produce simulation results pertaining to the COVID-19 pandemic available in a centralized database has expedited the trend of MD data sharing. Additionally, databases specifically designed to hold sets of simulations pertaining to membrane proteins, G-protein coupled receptors, and protein architectures have been constructed.
Due to file format heterogeneity and interoperability, a significant amount of MD simulation data remains unresolved at this time. Finding public data on particular proteins or parameters might be challenging because researchers frequently share their simulation files across numerous generic repositories. The dark matter surrounding MD is a serious problem that requires attention. It will be simpler to retrieve and reuse these files if this secret volume of data is unlocked. This will enhance the data’s fairness and reproducibility, improving the accessibility and fairness of MD simulation data.
About the Study
In this work, researchers have indexed scattered MD simulation files stored in generalist data repositories using a search approach. Researchers conducted a large-scale proof-of-concept analysis of publicly available MD data, concentrating on the files produced by the Gromacs MD software. Researchers demonstrated the many categories of the simulated molecules and the biophysical settings that were applied to these systems, as well as the high worth of these data. The study suggested a search engine prototype based on these findings and the annotations to investigate this MD dark matter quickly. Lastly, researchers offer straightforward recommendations for data sharing to progressively raise the FAIRness of MD data, building on this experience.
MDverse
For further research, the MD community has released files that it has gathered; comprehensive documentation is accessible on GitHub. Researchers can access datasets for all MD engines .gro and .mdp files with ease thanks to a prototype online application called MDverse Data Explorer. The program provides descriptions of the data it finds, allows users to search for keywords, and uses filters and keywords to obtain interesting datasets. A tab-separated values (.tsv) file containing the data can be exported for additional investigation.
Future Direction
Integrating data repositories such as Dryad and Dataverse instances, along with the collaborative source code platform GitHub, helps improve MD data analysis. Gromacs has 70,000 parameter .mdp files and 55,000 structure .gro files, which are short text-based MD simulation files that can be hosted on GitHub. The scripts that are included with these files can offer insightful information on MD analysis. By creating links to simulation data, GitHub repositories can also serve as access points to other datasets. Nevertheless, long-term availability is not guaranteed by repositories such as GitHub. Further improving accessibility and reusability of MD data can be achieved by enriching metadata and integrating it into the research environment.
Conclusion
Now that 250,000 files from 2,000 datasets have been indexed on sites like Zenodo, Figshare, and OSF, MD simulations are a widespread practice. If you are a researcher and cannot access HPC facilities or would prefer not to use expensive simulations, this trend provides opportunities and incentives. There are currently a large number of available simulations, which enable researchers to compare simulations of specialized molecular systems, extend simulations with equilibrated systems, and reanalyze previously completed trajectories. It is essential to build annotated, well-maintained datasets to create dynamic, generative deep-learning models for artificial intelligence. The reuse and reproducibility of MD simulations are enhanced by improving metadata and accessible data. This demonstrates the importance of setting up and producing MDs according to best practices.
Article Source: Reference Paper | MDverse Data Explorer: Web Application | The code of the MDverse data explorer web application is available freely on GitHub.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.