The Brain Language Model (BrainLM), a foundation model for brain activity dynamics trained on 6,700 hours of functional magnetic resonance imaging (fMRI) scans, was introduced by researchers at Yale University. Through self-supervised masked-prediction training, BrainLM exhibits high performance on zero-shot inference and fine-tuning tasks.
Accurate prediction of future brain states and clinical variables such as age, anxiety, and PTSD are made possible by fine-tuning. Importantly, the BrainLM performance was admirable when applied to completely fresh external cohorts that were not present during training. BrainLM does not require network-based supervision during training to identify intrinsic functional networks straight from raw fMRI data while operating in zero-shot inference mode. Additionally, the model produces interpretable latent representations that show connections between patterns of brain activity and states of cognition. BrainLM provides a flexible and understandable framework for clarifying the intricate spatiotemporal dynamics of brain activity in humans. It acts as a potent “lens” that allows large-scale fMRI data banks to be analyzed in novel ways, leading to more efficient interpretation and application. The research shows how foundation models can be used to further computational neuroscience studies.
Understanding how brain activity drives cognition and behavior requires an understanding of blood-oxygen-level dependent (BOLD) signals, an indirect indicator of brain function. By monitoring changes in blood oxygen levels that correspond to specific neuronal activity in a given area, functional magnetic resonance imaging provides a noninvasive window into the functioning brain. However, because of their complicated spatiotemporal dynamics and relationships across space and time, analyzing the vast, high-dimensional recordings presents significant hurdles. These nonlinear interactions are not well-modeled by the majority of current analysis techniques. Previous methods of fMRI analysis have placed a great deal of reliance on machine learning models, which has limited their integration and generalizability to unlabeled fMRI data. As a result, adaptable modeling frameworks are required to properly utilize the size and complexity of fMRI repositories.
A new paradigm in artificial intelligence is represented by foundation models, which replace limited, task-specific training with more flexible and universal models. The foundation model technique, which is motivated by advances in natural language processing, trains adaptable models at scale on a large amount of data, opening up a wide range of downstream capabilities through transfer learning. In contrast to earlier AI systems created for specific tasks, foundation models provide broad computational capabilities that render them appropriate for a vast range of real-world uses. GPT and other large language models have shown the framework’s potential in a variety of fields, such as education, healthcare, robotics, and more. New perspectives on problems in neuroscience and medical imaging analysis are provided by foundation models.
What is BrainLM?
BrainLM is the first fMRI recording foundation model. The Transformer-based design used by BrainLM allows it to capture the spatiotemporal dynamics seen in large-scale brain activity data. Unsupervised representation learning without task-specific constraints is made possible through pretraining on a substantial corpus of raw fMRI recordings. Following pretraining, BrainLM uses zero-shot inference and fine-tuning to serve a variety of downstream applications. The researchers used important tasks such as future brain state prediction, cognitive variable decoding, and functional network identification to showcase BrainLM’s potential. The collective outcomes demonstrate BrainLM’s ability to perform well on tasks involving both zero-shot and fine-tuning. This paper demonstrates how large language models can be used to further neuroscience research. By offering more potent computational tools to clarify the complex workings of the human brain, BrainLM serves as a foundation model on which the community can expand.
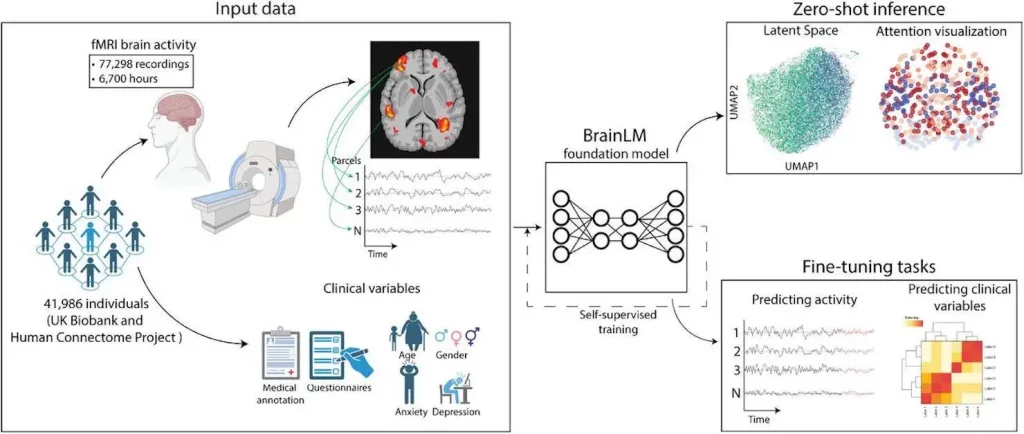
Image Source: https://doi.org/10.1101/2023.09.12.557460
BrainLM’s Abilities
- Model Generalization: By demonstrating strong performance on held-out recordings from both the independent HCP dataset and the UK Biobank (UKB) test set, the machine learning model BrainLM proved its capacity to generalize to fresh fMRI data. Strong generalization on previously unseen recordings from the same distribution was indicated by the model’s average R2 score of 0.464 on the UKB test data. BrainLM accurately captured the dynamics of the brain in this new distribution, even with variations in cohort and acquisition specifics. One can see the benefits of large-scale pretraining for learning broadly applicable models of brain activity from the model generalization, which is scaled with data size and model size.
- Prediction of Clinical Variables: BrainLM is a pre-trained model designed to represent clinical factors associated with fMRI measurements precisely. The prediction power of the model has been improved by fine-tuning it to regress metadata factors from the UKB dataset. With a lower mean squared error than competing techniques, the model consistently performed better than alternative approaches. By evaluating their zero-shot metadata regression performance, the models’ recorded embeddings were evaluated for quality. Larger models gain pretraining knowledge of more therapeutically relevant data. BrainLM provides a strong foundation for fMRI-based evaluation of cognitive health and brain diseases. These results demonstrate BrainLM’s capacity to identify predictive signals within intricate fMRI recordings. Clinical prediction is being investigated in a wider range of neurological, neurodegenerative, and psychiatric diseases.
- Prediction of Future Brain States: When projecting to future brain states using UKB data, BrainLM— used to anticipate parcel activity at future time points—performed better than expected. After being trained to predict 20 time steps, the model was refined using 180 time-step sequences. In contrast to baseline models like LSTMs, NODE, and an untrained version, the optimized BrainLM model consistently recorded the lowest error throughout all projected time-steps, proving its strong capacity to intuitively understand the fMRI dynamics.
- Interpretability via Attention Analysis: The interpretability of BrainLM is a critical aspect that can be improved by examining self-attention weights. A deeper understanding of the internal representations of the model is therefore made possible. Attention patterns were noted during fMRI recordings, with a clear emphasis on the visual cortex. The frontal and limbic brain regions were prominently highlighted in patients with severe depression, which is consistent with functional changes linked to depression. These attention maps demonstrate how BrainLM may identify changes in functional networks that are clinically relevant, enhancing interpretive abilities and offering insightful information about neuroscience.
- Functional Network Prediction: Without network-based supervision, BrainLM’s capacity to divide parcels into functional brain networks was assessed. The study benchmarked different approaches for sorting parcels into these networks and categorized them into seven functional categories. BrainLM’s attention-driven method outperformed previous approaches, with 58.8% accuracy in parcel classification. With only 25.9% accuracy, the GCN-based k-NN classifier trailed behind. These results demonstrate how well BrainLM can identify functional brain topography even in the pretraining stage, with self-attention maps offering significant information about the identification of the network.
Conclusion
BrainLM is an innovative foundation model that predicts and interprets human brain dynamics using self-supervised pre-training on 6,700 hours of recorded brain activity. Reconstructing masked brain activity sequences with good accuracy becomes better with more factors, indicating the advantages of extensive pretraining. Additionally, BrainLM offers a strong foundation for the development of biomarkers, which improves the prediction of clinical factors and mental illnesses. After pretraining, it may immediately identify intrinsic functional connection maps by clustering parcels into systems that are already known. Future research may combine fMRI with other brain-related data, such as structural, functional, and genetic information, or with other recording modalities like EEG and MEG. BrainLM is a platform enabling faster research at the nexus of AI and neuroscience to clarify the complex functions of the human brain.
Article Source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.