A typical gigapixel slide may include tens of thousands of picture tiles, which presents special computing issues for digital pathology. The crucial slide-level context has been lost in previous models, which frequently resorted to subsampling a small portion of tiles for each presentation. The researchers introduce Prov-GigaPath, a foundation model for whole-slide pathology that was pretrained on 1.3 billion 256 × 256 pathology picture tiles in 171,189 entire slides from Providence, a sizable US health network that includes 28 cancer facilities. The slides came from almost 30,000 patients, representing 31 main tissue categories. Researchers suggest GigaPath, a unique vision transformer architecture for pretraining gigapixel pathology slides, as a way to train Prov-GigaPath. A slide-level learning model, GigaPath, uses image tiles to perform problems related to digital pathology. It modifies the LongNet technique to manage thousands of tasks. On 25 tasks out of 26, the open-weight foundation model Prov-GigaPath performs better than the second-best approach, with notable gains on 18 workloads. Including pathology data also demonstrates potential in vision-language pretraining for pathology. This model underlines how crucial whole-slide modeling and real-world data are for digital pathology jobs.
Introduction
In computational pathology, which has the potential to revolutionize cancer diagnosis, cancer staging and diagnostic prediction are essential applications. Nevertheless, current computational methods frequently demand substantial volumes of annotated data and are designed for certain purposes. By using unlabeled data to pretrain foundation models, self-supervised learning has demonstrated encouraging results in lowering task-specific annotations. In order to alleviate the data annotation bottleneck in computational pathology, these models have been designed for biological areas where labeled data is rare but unlabelled data is abundant.
Challenges Hinder the Development and Use of Pathology Foundation Models for Real-World Clinical Applications
- Pathology models are pretrained using the extensive data set of the Cancer Genome Atlas (TCGA). The efficacy of these models is, however, constrained by the poor quality and scarcity of this data. Performance decreases may occur when applying TCGA-based predictive models and biomarkers on out-of-distribution samples because the TCGA data, which comprises 30,000 slides and 208 million image tiles, might not be enough to handle real-world digital pathology issues like heterogeneity and noise artifacts.
- The hierarchical model architecture of the Hierarchical Image Pyramid Transformer (HIPT) is intended to capture global patterns spanning whole slides as well as local patterns inside individual tiles. HIPT employs hierarchical self-attention over tiles, which differs from previous models and enables more intricate global patterns in entire gigapixel slides.
- Third, the foundation models produced by the rare instances in which pretraining has been carried out on extensive real-world patient data are usually unavailable to the general public, restricting their wider applicability in clinical research and applications.
Introducing Prov-GigaPath
The Providence Health Network created the open-weight pathology foundation concept known as Prov-GigaPath. A sizable digital pathology dataset, comprising 1,384,860,229 picture tiles from 171,189 pathology slides from more than 30,000 patients, is used for pretrained training. In terms of picture tiles, this dataset is more than five times larger than the TCGA, and in terms of patients, it is twice as enormous. Prov-GigaPath is the largest pretraining effort to date, containing essential information such as histopathological findings, cancer staging, genetic mutation profiles, and related pathology reports.
Understanding GigaPath
GigaPath is a vision transformer created by researchers from Providence Health System, the University of Washington’s Paul G. Allen School of Computer Science & Engineering, and Microsoft, which is used to pretrain massive pathology foundation models using gigapixel pathology slides. A slide becomes a lengthy series of tokens by embedding image tiles as visual tokens using this creative method. However, because there are so many tiles on a slide, traditional vision converters cannot be applied directly to digital pathology. To tackle this, the researchers created LongNet, a dilated self-attention technique that combines masked autoencoder with LongNet to enable whole-slide self-supervised learning after first enabling image-level self-supervised learning using DINOv2.
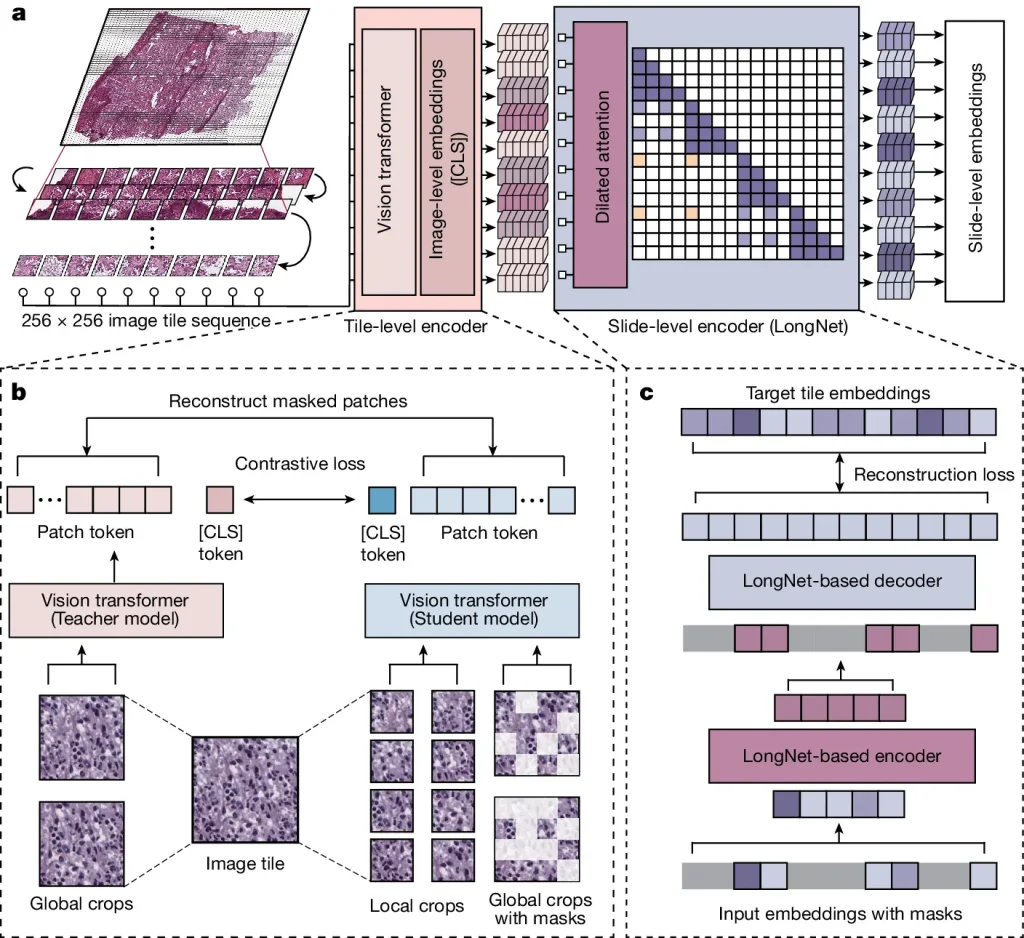
Image Source: https://doi.org/10.1038/s41586-024-07441-w
Methodology of Prov-GigaPath
An in-depth analysis and practical evaluation of the Prov-GigaPath digital pathology foundation concept have been conducted. Concerning cutting-edge models such as HIPT, CtransPath, and REMEDIS, the fully open-weight model has demonstrated a notable performance gain. In six cancer types, Prov-GigaPath significantly exceeds the second-best strategy, and it outperforms all other models in all nine forms of cancer. Its large-scale pretraining and extremely large-context modeling contribute to its broad applicability across cancer types. The model demonstrates its potential for multimodal integrative data analysis by displaying state-of-the-art performance in standard vision-language modeling tasks, such as mutation prediction and zero-shot subtyping. Large-scale machine learning models in the realm of digital pathology are used in this study to show how Prov-GigaPath may enhance clinical diagnostics and decision support procedures.
Conclusion
A foundation model for pathology known as Prov-GigaPath was created with a variety of digital pathology applications in mind. Prov-GigaPath allows gigapixel WSN modeling at an ultra-large context, having been pre-trained on a sizable real-world dataset from Providence Health System. The model, which was developed by modifying LongNet as the vision transformer, showed cutting-edge results on tasks related to pathomics, cancer subtyping, and vision-language processing. Prov-GigaPath can be useful for more general biomedical domains for effective self-supervised learning from high-resolution images. It can also help with clinical diagnostics and decision support.
To learn more about GigaPath, kindly refer to the Reference Paper and Reference Article. Prov-GigaPath can be accessed at GitHub.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.