Research on genomes is booming in the scientific community, and this is translating into an explosion of published results. Genetic experts and researchers now need to keep up with the most recent discoveries and their practical implications. Finding pertinent scientific articles and gathering important data for the interpretation of genetic variants provide a problem. The process of interpreting genetic variants can be made more effective by using computer-aided literature search and summarization to convert large genomic datasets into clinically applicable insights. In order to close this gap, researchers from the University of Pavia introduced VarChat (varchat.engenome.com), a cutting-edge application built on generative AI that was created to locate and condense the dispersed scientific literature related to genetic variants into concise but educational writings. VarChat gives users a succinct explanation of particular genetic variants, including information on how they affect linked proteins and potential health implications for humans. VarChat additionally promotes further research and provides direct links to relevant, reliable scientific sources.
Introduction
Genomics and personalized medicine are producing massive amounts of data, with genetic variations being the main area of study. The great majority of these variations are well-documented in scientific publications, and they can be connected to treatment responses, disease susceptibility, and other phenotypic outcomes. In addition to the detection of these variants from sequencing data, there have been various proposals for automating the process of effectively curating and interpreting this vast amount of data. Much work has gone into compiling this knowledge into publicly accessible resources and specialized databases such as gnomAD, OMIM, and ClinVar. With its extensive body of knowledge, the scientific literature provides a multitude of insights into these genetic variations. However, manual curation is difficult due to the massive volume of publications and the lack of precise standardization in the use of a nomenclature characterizing genetic variations. Additionally, the curation process—which is much enhanced by having access to complete texts and supplemental data of scientific articles—is a crucial component in improving the finding of genetic variants.
Tools to Support Genomic Variant Research
LitVar is a semantic search engine created especially to connect PMC and PubMed genetic variant data. Advanced text mining algorithms are used by LitVar to visualize the relationships between variations and other connected entities, such as diseases and chemicals/drugs, in addition to retrieving standardized variant information. Another tool that aims to assist in the curation of genomic variants is called Variomes, which is a high-recall search engine. By changing the parameters, one can add keywords for articles to be reranked and customize the search by defining the chronology. Lastly, SynVar, which offers descriptions in both standard and non-standard formats common in the literature, has been developed to guarantee efficient retrieval of texts containing variants.
The incapacity of these methods to combine the information from variants into succinct, accessible paragraphs appropriate for clinical reports is a serious drawback. While many systems focus on data aggregation and classification, they are not as successful in producing thorough yet concise textual interpretations.
Looking Into Large Language Models
Generative AI-based large language models (LLMs), such as the well-known chatGPT (www.openai.com), Bard (google AI), Falcon (www.tii.ae), and Claude 2 (Anthropic, www.claude.ai), are naturally capable of understanding and summarising complicated texts. These models are built on a deep learning architecture called a Transformer, which has millions or billions of parameters and an inventive “attention” layer that allows the model to assign varying weights to different input components, enhancing its comprehension of the context and relationships in the data. They have proven to perform exceptionally well on a variety of Natural Language Processing tasks, exhibiting great understanding and reasoning skills, and have become essential components of solutions that are widely utilized in our daily lives.
Introducing VarChat
It is still difficult to strike a balance between offering precise and in-depth genetic insights and making sure that the writing is understandable to a wide range of readers, including those who lack extensive computational and genomic knowledge.
For this reason, researchers present VarChat, the first generative AI-based application created to find and compile scientific literature regarding a variant in a human genome, together with a brief description of the variant, key findings from previous studies, and related references.
Methodology
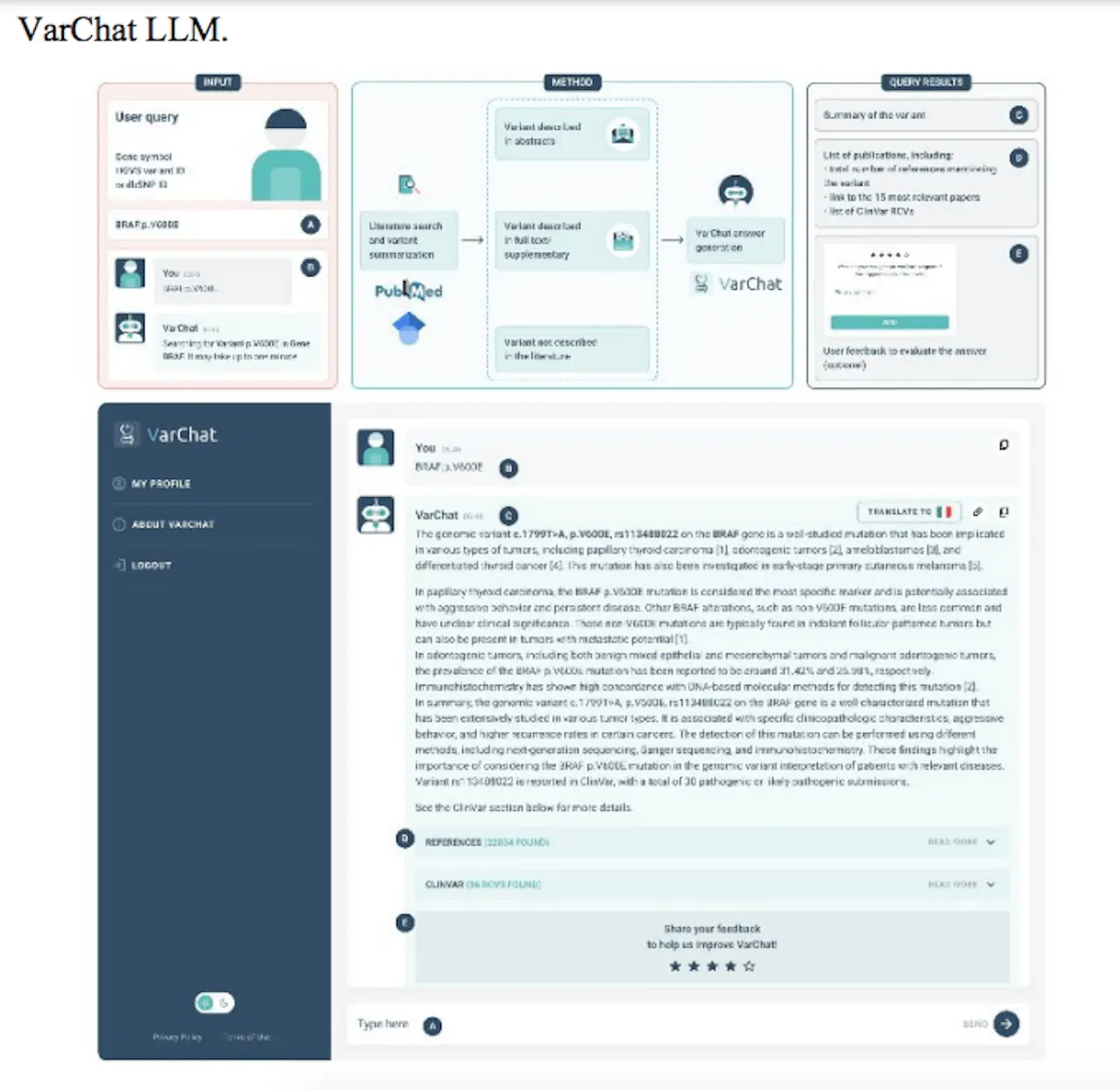
VarChat needs the dbSNP identifier or the genomic variant coordinates in accordance with the HGVS nomenclature as input. Through the use of an LLM model, VarChat generates succinct and logical summaries for each variant that is queried, allowing researchers and clinicians to grasp the key points of the publications related to these variants. When available, the system furnishes the user with the top 15 pertinent references along with a written summary. A modified version of the BM25 ranking algorithm, which mostly uses the classic word frequency, is used to determine how relevant the publication is. Papers that report the variant only in the supplementary are penalized; more weight is given to those that cite the variant in the abstract and were published within the last two years.
In addition, if the variant is found in ClinVar, a direct link to the database page, the related records, the condition that is linked to it, its clinical importance, and the review status are shown. Last but not least, the ability to translate the response into thirty different languages has been activated. While the Restful API is based on a serverless and scalable infrastructure that makes use of Python 3, FastAPI, and Amazon AWS Lambda functions, it is designed to be both desktop and mobile-optimized. VarChat’s graphical user interface works with ReactJS.
Application of VarChat
Using the HGVS nomenclature, users can search for genomic variants by selecting a gene symbol along with any or both of the following: coding DNA reference sequence, protein reference sequence, mitochondrial DNA reference sequence, or even both. “BRAF:p.V600E,” “PINK1:c.926G>A,” “MT-ND4:m.11778G>A,” and “rs34637584” are a few examples of legitimate searches. Researchers advise utilizing mitochondrial coordinates for mitochondrial variations, such as “MT-ND4:m.11778G>A” as an example.
Genomic coordinates are not supported as input at this time. Each variant’s HGVS or rs dbSNP ID can be determined using freely available annotation and interpretation software like VEP or conversion tools like TransVar. Researchers retrieve all scientific papers that reference the variant in the full text, abstract, or supplemental information.
The user is presented with the fifteen most pertinent papers and direct links to the full-text publications. VarChat then uses the abstracts associated with the sought variant for summarization.
VarChat uses the available data to produce a summary based on insights from its LLM model and pertinent papers.
In particular, the text references the variant as supplementary content for the summarization if it appears in the PubMed abstract or the freely accessible Google Scholar papers. If not, the VarChat LLM generates the response in its entirety. In any case, the list of references for the variant that relates to it is displayed if there is a variant that matches the scientific literature.
Users are meant to be able to trust the system. VarChat, being an LLM-based system, has the potential to cause “hallucinations,” a situation in which these models produce information that is factually wrong or not supported by the input data. When LLMs are used for activities requiring high levels of accuracy and reliability, this aspect might be very difficult.
VarChat makes it very evident to users where its answers come from, letting them know if they were created using only VarChat LLM’s expertise or from references in an effort to increase process transparency.
Conclusion
VarChat is the first generative AI tool created expressly to aid with the interpretation of genomic variants. It does this by quickly locating and synthesizing pertinent scientific literature, thus serving as a genetic assistant. VarChat has the potential to be a useful tool for the genetics community in evaluating human genetic variations through generative AI, improving knowledge of the implications and effects of variants.
Article source: Reference Paper | VarChat can be accessed at varchat.engenome.com at no cost.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.